Genetic and physical interactions involving the yeast nuclear cap-binding complex - PubMed (original) (raw)
Genetic and physical interactions involving the yeast nuclear cap-binding complex
P Fortes et al. Mol Cell Biol. 1999 Oct.
Abstract
Yeast strains lacking the yeast nuclear cap-binding complex (yCBC) are viable, although impaired in growth. We have taken advantage of this observation to carry out a genetic screen for components that show synthetic lethality (SL) with a cbp20-Delta cbp80-Delta double mutation. One set of SL interactions was due to mutations that were complemented by components of U1 small nuclear RNP (snRNP) and the yeast splicing commitment complex. These interactions confirm the role of yCBC in commitment complex formation. Physical interaction of yCBC with the commitment complex components Mud10p and Mud2p, which may directly mediate yCBC function, was demonstrated. Unexpectedly, we identified multiple SL mutations that were complemented by Cbf5p and Nop58p. These are components of the two major classes of yeast small nucleolar RNPs, which function in the maturation of rRNA precursors. Mutants lacking yCBC were found to be defective in rRNA processing. Analysis of the yCBC deletion phenotype suggests that this is likely to be due to a defect in the splicing of a subset of ribosomal protein mRNA precursors.
Figures
FIG. 1
yCBC affects vegetative growth rate. Strain YJV159 (wild type) was disrupted for GCR3 (cbp80-Δ), for MUD13 (cbp20-Δ), or for both (cbp20/80-Δ). These strains were grown to mid-log phase, and four dilutions of each were plated to compare the growth rates. The doubling time of each strain was also assayed in liquid culture and is indicated on the right.
FIG. 2
yCBC interacts with Mud2p and with Mud10p. [35S]methionine-labeled Mud2p/Luc2p (lanes 1 to 5), Snu71p/Luc5p (lanes 6 to 10), and Mud10p/Luc4p (lanes 11 to 15) were incubated with a control column (MOCK) or with a yCBC column as indicated. Samples were fractionated into nonbound supernatant (S) and bound pellet (P) fractions and analyzed by SDS-polyacrylamide gel electrophoresis. In lanes 1, 6, and 11, 25% of the input protein was loaded.
FIG. 3
Growth of the LUC8 and LUC9 strains carrying functional CBC. Dilutions (1- to 102-fold) of luc8 and luc9 strains along with the wild-type isogenic (WT) control strain were spotted on minimal plates at 16, 23, 30, and 37°C and incubated for 3 days.
FIG. 4
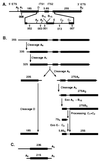
The yeast pre-rRNA processing pathway. (A) Structure of the pre-rRNA with positions of oligonucleotides used for hybridization. In the 35S pre-rRNA, the mature 18S, 5.8S, and 25S rRNA sequences are flanked by the 5′ and 3′ external transcribed spacers (5′ ETS and 3′ ETS) and separated by internal transcribed spacers 1 and 2 (ITS1 and ITS2). (B) Major pre-rRNA processing pathway in yeast. Note that a minor alternative pathway in ITS1 generates an alternative form of 5.8S rRNA (5.8SL) that is extended 5′ to site BIL (not shown). (C) Structures of the aberrant 23S and 21S RNAs.
FIG. 5
Northern analysis of pre-rRNA (A and C) and snoRNA (B and D) levels in LUC8 and LUC9 strains. RNA was extracted following growth at 23°C and 18 h after transfer to 37°C (A and B) or following growth at 30°C and 12 h after transfer to 16°C (C and D). The oligonucleotide probes used in panels A and C were 003 (top) and 002 (bottom). WT, wild type.
FIG. 6
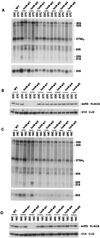
yCBC is required for normal pre-rRNA processing. For Northern blot analysis of mature and precursor rRNAs, RNA was extracted from wild-type (WT) and cbp strains as indicated. (A) Hybridization with a probe complementary to the mature 18S and 25S RNAs; (B) hybridization with a probe complementary to the 5′ region of ITS1 (oligonucleotide 002); (C) hybridization with a probe complementary to the 3′ region of ITS1, downstream of site A3 (oligonucleotide 001); (D) hybridization with a probe complementary to the central region of ITS1, between sites A2 and A3 (oligonucleotide 003); (E and F) hybridization with a probe complementary to the 5′ region of ITS2 (oligonucleotide 013). (G) Ratios of steady-state levels of mature 18S and 25S rRNAs. The positions of mature and precursor rRNA species are indicated; 21S and 20S pre-rRNAs are not well resolved; the identity of the 32S intermediate was verified by hybridizing a riboprobe complementary to the region between sites A0 and A1. Positions of the oligonucleotide probes are depicted in Fig. 4.
FIG. 7
yCBC does not affect accumulation of various snoRNAs. For Northern blot analysis of low-molecular-weight RNAs, RNA was extracted from wild-type and cbp strains as indicated. The probes used for hybridization are described in Materials and Methods.
FIG. 8
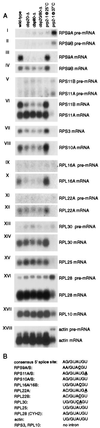
yCBC affects steady-state levels of mRNAs of ribosomal proteins. RNA was extracted from either wild-type or cbp strains as indicated. Additionally, control RNA was extracted from a temperature-sensitive-lethal splicing-deficient prp2-1 strain grown either at the permissive temperature (25°C) or after shift to the nonpermissive temperature (37°C) for 60 min. (A) Analysis of RPS and RPL mRNAs and pre-mRNAs. The positions of mature and precursor mRNAs are indicated. Probes used for hybridization are described in Materials and Methods. (B) Sequences at the 5′ splice sites of pre-mRNAs analyzed in this study. Nonconsensus residues are underlined. All pre-mRNAs tested contain one intron, and in all cases it is located close to the 5′ end of the pre-mRNA.
Similar articles
- The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex.
Colot HV, Stutz F, Rosbash M. Colot HV, et al. Genes Dev. 1996 Jul 1;10(13):1699-708. doi: 10.1101/gad.10.13.1699. Genes Dev. 1996. PMID: 8682299 - A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing.
Lewis JD, Görlich D, Mattaj IW. Lewis JD, et al. Nucleic Acids Res. 1996 Sep 1;24(17):3332-6. doi: 10.1093/nar/24.17.3332. Nucleic Acids Res. 1996. PMID: 8811086 Free PMC article. - Genetic interactions of hypomorphic mutations in the m7G cap-binding pocket of yeast nuclear cap binding complex: an essential role for Cbc2 in meiosis via splicing of MER3 pre-mRNA.
Qiu ZR, Chico L, Chang J, Shuman S, Schwer B. Qiu ZR, et al. RNA. 2012 Nov;18(11):1996-2011. doi: 10.1261/rna.033746.112. Epub 2012 Sep 21. RNA. 2012. PMID: 23002122 Free PMC article. - The cap binding complex influences H2B ubiquitination by facilitating splicing of the SUS1 pre-mRNA.
Hossain MA, Claggett JM, Nguyen T, Johnson TL. Hossain MA, et al. RNA. 2009 Aug;15(8):1515-27. doi: 10.1261/rna.1540409. Epub 2009 Jun 26. RNA. 2009. PMID: 19561118 Free PMC article. - Luc7p, a novel yeast U1 snRNP protein with a role in 5' splice site recognition.
Fortes P, Bilbao-Cortés D, Fornerod M, Rigaut G, Raymond W, Séraphin B, Mattaj IW. Fortes P, et al. Genes Dev. 1999 Sep 15;13(18):2425-38. doi: 10.1101/gad.13.18.2425. Genes Dev. 1999. PMID: 10500099 Free PMC article.
Cited by
- The distribution of active RNA polymerase II along the transcribed region is gene-specific and controlled by elongation factors.
Rodríguez-Gil A, García-Martínez J, Pelechano V, Muñoz-Centeno Mde L, Geli V, Pérez-Ortín JE, Chávez S. Rodríguez-Gil A, et al. Nucleic Acids Res. 2010 Aug;38(14):4651-64. doi: 10.1093/nar/gkq215. Epub 2010 Apr 12. Nucleic Acids Res. 2010. PMID: 20385590 Free PMC article. - The interaction of the cap-binding complex (CBC) with eIF4G is dispensable for translation in yeast.
Baron-Benhamou J, Fortes P, Inada T, Preiss T, Hentze MW. Baron-Benhamou J, et al. RNA. 2003 Jun;9(6):654-62. doi: 10.1261/rna.5100903. RNA. 2003. PMID: 12756324 Free PMC article. - A mutation in the Cap Binding Protein 20 gene confers drought tolerance to Arabidopsis.
Papp I, Mur LA, Dalmadi A, Dulai S, Koncz C. Papp I, et al. Plant Mol Biol. 2004 Jul;55(5):679-86. doi: 10.1007/s11103-004-1680-2. Plant Mol Biol. 2004. PMID: 15604709 - Domain Requirements and Genetic Interactions of the Mud1 Subunit of the Saccharomyces cerevisiae U1 snRNP.
Agarwal R, Schwer B, Shuman S. Agarwal R, et al. G3 (Bethesda). 2019 Jan 9;9(1):145-151. doi: 10.1534/g3.118.200781. G3 (Bethesda). 2019. PMID: 30413416 Free PMC article. - Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae.
Das B, Butler JS, Sherman F. Das B, et al. Mol Cell Biol. 2003 Aug;23(16):5502-15. doi: 10.1128/MCB.23.16.5502-5515.2003. Mol Cell Biol. 2003. PMID: 12897126 Free PMC article.
References
- Abovich N, Liao X C, Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8:843–854. - PubMed
- Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. - PubMed
- Baudin-Baillieu A, Guillemet E, Cullinand C, Lacroute F. Construction of a yeast strain deleted for the TRP1 promoter and coding region that enhances the efficiency of the polymerase chain reaction-disruption method. Yeast. 1997;13:353–356. - PubMed
- Berben G, Dumont J, Gilliquet V, Bolle P A, Hilger F. The Ydp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous