The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae - PubMed (original) (raw)
The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae
A Eberharter et al. Mol Cell Biol. 1999 Oct.
Abstract
We have identified two Gcn5-dependent histone acetyltransferase (HAT) complexes from Saccharomyces cerevisiae, the 0.8-MDa ADA complex and the 1.8-MDa SAGA complex. The SAGA (Spt-Ada-Gcn5-acetyltransferase) complex contains several subunits which also function as part of other protein complexes, including a subset of TATA box binding protein-associated factors (TAFIIs) and Tra1. These observations raise the question of whether the 0.8-MDa ADA complex is a subcomplex of SAGA or whether it is a distinct HAT complex that also shares subunits with SAGA. To address this issue, we sought to determine if the ADA complex contained subunits that are not present in the SAGA complex. In this study, we report the purification of the ADA complex over 10 chromatographic steps. By a combination of mass spectrometry analysis and immunoblotting, we demonstrate that the adapter proteins Ada2, Ada3, and Gcn5 are indeed integral components of ADA. Furthermore, we identify the product of the S. cerevisiae gene YOR023C as a novel subunit of the ADA complex and name it Ahc1 for ADA HAT complex component 1. Biochemical functions of YOR023C have not been reported. However, AHC1 in high copy numbers suppresses the cold sensitivity caused by particular mutations in HTA1 (I. Pinto and F. Winston, personal communication), which encodes histone H2A (J. N. Hirschhorn et al., Mol. Cell. Biol. 15:1999-2009, 1995). Deletion of AHC1 disrupted the integrity of the ADA complex but did not affect SAGA or give rise to classic Ada(-) phenotypes. These results indicate that Gcn5, Ada2, and Ada3 function as part of a unique HAT complex (ADA) and represent shared subunits between this complex and SAGA.
Figures
FIG. 1
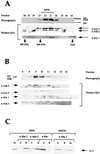
Purification of ADA. (A) Schematic representation of the chromatographic steps applied for purification of the ADA complex. ADA was followed by HAT assay and immunoblotting. (B) Silver staining of purified ADA. Aliquots of ADA peak fractions which were eluted from the last three chromatographic steps (Superose 6, Mini Q, and Superose 6 PC 3.2/30) were separated by SDS-polyacrylamide gel electrophoresis and stained with silver. For the final Superose 6 column, side fractions are also shown. Arrows and asterisks indicate proteins which were coeluted with the purified ADA fraction in amounts apparently stoichiometric with one another.
FIG. 2
Mass spectrometry analysis. Mass spectrometry identified the four ADA subunits as Ada2, Ada3, Gcn5, and Ahc1. (A) Peptide sequences obtained by mass spectrometry for Ada2, Ada3, Gcn5, and Ahc1. Numbers at the left and the right of each sequence indicate the first and the last amino acids identified, respectively. For all four proteins, numerous hits were obtained. (B) Complete amino acid sequence for Ahc1. The underlined 16 N-terminal amino acid residues were used to generate the Ahc1 antiserum. Residues shown in boldface represent amino acids identified by mass spectrometry.
FIG. 3
Western blot analysis of Mono Q chromatography. Ten microliters of indicated fractions was separated after Mono Q chromatography by SDS–10% polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes. Membranes were incubated with antibodies (α) against Ahc1, Ada2, Ada3, and Spt8.
FIG. 4
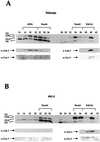
Ahc1 is coeluted with purified ADA. (A) Fractions from the seventh column, Superose 6 size exclusion chromatography, were tested in a nucleosomal HAT assay and Western blotting with the indicated antibodies. The upper panel shows the fluorogram from the HAT assay depicting the specificity of ADA. Histones H3 and H2B were acetylated by ADA. (B) Fluorogram after nucleosomal HAT assay and Western blotting with the indicated antibodies (α) of purified fractions from the Mini Q column. (C) Immunoprecipitation with purified ADA and SAGA complex. ADA and SAGA were incubated with preimmune serum, anti-Ada2 antiserum, or anti-Ahc1 antiserum immobilized on protein A-Sepharose beads. The fluorogram of HAT reaction products obtained with free histones is shown.
FIG. 5
An AHC1 mutant specifically affects the ADA HAT complex. Whole-cell extracts from a wild-type strain and a yeast strain bearing a mutation in AHC1 were partially purified with Ni2+ NTA agarose and Mono Q chromatography. (A) In the top panel, a typical fluorogram from a nucleosomal HAT assay with fractions from a Mono Q column prepared from a wild-type strain is presented. The four HAT complexes are indicated at the top. The lower panels show results from Western blotting with antibodies (α) raised against Ada2 and Gcn5. (B) Fluorogram from a nucleosomal HAT assay of Mono Q fractions prepared from the _ahc1_Δ yeast strain. NuA4, NuA3, and SAGA were eluted in the same fractions as in the wild type. ADA was absent in the AHC1 mutant. Western blots (lower panels) demonstrate that anti-Ada2 and anti-Gcn5 antibodies showed immunoreactivity only for SAGA (fractions 38 to 40).
FIG. 6
The ADA HAT complex can be rescued by plasmid expression of AHC1. (A and B) Mono Q fractionation of partially purified whole-cell extracts prepared from YJW 103 (_ahc1_Δ) and YJW 104 (_ahc1_Δ-pAHC1:HA3). HAT assay fluorograms and Western blots of fractions containing ADA and NuA4 (fractions 14 to 24) are shown. Immunodetection of Ahc1 was accomplished with an anti-HA antibody. The ADA HAT complex was specifically restored in YJW 104 (B) and was absent in the AHC1 deletion (A). HAT assays (C) and Western blots (D) from Superose 6 size exclusion chromatography of Mono Q fractions (A) are shown. Fractions 14 to 20 from a Mono Q column were pooled, concentrated, and fractionated on a Superose 6 column. Ahc1 was immunodetected by monoclonal anti-HA antibody. ADA is present only in YJW 104 bearing pAHC1-HA3 and was eluted at a molecular mass of ∼800 kDa.
FIG. 7
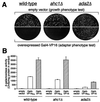
An AHC1 deletion does not display a classic Ada phenotype. (A) Transformants of wild-type, _ahc1_Δ, and _ada2_Δ (adapter control) cells containing high-copy-number empty vector were plated on minimal medium to assess overall growth phenotype (upper panels). To test for adapter phenotype (Ada−; relief of toxicity of overexpressed chimeric activator Gal4-VP16), cells were transformed with high-copy-number activator plasmid and plated on minimal medium (lower panels). (B) Quantitation of acidic-activator-mediated in vivo transcription is presented. Wild-type, _ahc1_Δ, and _ada2_Δ cells were transformed with pLGSD5 reporter plasmid and low-copy-number empty vector, Gal4-VP16, or Gal4-VP16FA plasmids, and extracts were assayed for β-galactosidase activity.
Similar articles
- Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex.
Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. Grant PA, et al. Genes Dev. 1997 Jul 1;11(13):1640-50. doi: 10.1101/gad.11.13.1640. Genes Dev. 1997. PMID: 9224714 - Adenovirus E1A requires the yeast SAGA histone acetyltransferase complex and associates with SAGA components Gcn5 and Tra1.
Kulesza CA, Van Buskirk HA, Cole MD, Reese JC, Smith MM, Engel DA. Kulesza CA, et al. Oncogene. 2002 Feb 21;21(9):1411-22. doi: 10.1038/sj.onc.1205201. Oncogene. 2002. PMID: 11857084 - The SAGA HAT module is tethered by its SWIRM domain and modulates activity of the SAGA DUB module.
Haile ST, Rahman S, Fields JK, Orsburn BC, Bumpus NN, Wolberger C. Haile ST, et al. Biochim Biophys Acta Gene Regul Mech. 2023 Jun;1866(2):194929. doi: 10.1016/j.bbagrm.2023.194929. Epub 2023 Mar 24. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 36965704 Free PMC article. - Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain.
Wallberg AE, Flinn EM, Gustafsson JA, Wright AP. Wallberg AE, et al. Biochem Soc Trans. 2000;28(4):410-4. Biochem Soc Trans. 2000. PMID: 10961930 Review. - The Ada2/Ada3/Gcn5/Sgf29 histone acetyltransferase module.
Espinola-Lopez JM, Tan S. Espinola-Lopez JM, et al. Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194629. doi: 10.1016/j.bbagrm.2020.194629. Epub 2020 Sep 2. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32890768 Free PMC article. Review.
Cited by
- An integrated SAGA and TFIID PIC assembly pathway selective for poised and induced promoters.
Mittal C, Lang O, Lai WKM, Pugh BF. Mittal C, et al. Genes Dev. 2022 Sep 1;36(17-18):985-1001. doi: 10.1101/gad.350026.122. Epub 2022 Oct 27. Genes Dev. 2022. PMID: 36302553 Free PMC article. - Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8.
Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Henry KW, et al. Genes Dev. 2003 Nov 1;17(21):2648-63. doi: 10.1101/gad.1144003. Epub 2003 Oct 16. Genes Dev. 2003. PMID: 14563679 Free PMC article. - A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA.
Friis RM, Wu BP, Reinke SN, Hockman DJ, Sykes BD, Schultz MC. Friis RM, et al. Nucleic Acids Res. 2009 Jul;37(12):3969-80. doi: 10.1093/nar/gkp270. Epub 2009 Apr 30. Nucleic Acids Res. 2009. PMID: 19406923 Free PMC article. - The bromodomain of Gcn5 regulates site specificity of lysine acetylation on histone H3.
Cieniewicz AM, Moreland L, Ringel AE, Mackintosh SG, Raman A, Gilbert TM, Wolberger C, Tackett AJ, Taverna SD. Cieniewicz AM, et al. Mol Cell Proteomics. 2014 Nov;13(11):2896-910. doi: 10.1074/mcp.M114.038174. Epub 2014 Aug 8. Mol Cell Proteomics. 2014. PMID: 25106422 Free PMC article. - Gcn5-mediated Rph1 acetylation regulates its autophagic degradation under DNA damage stress.
Li F, Zheng LD, Chen X, Zhao X, Briggs SD, Du HN. Li F, et al. Nucleic Acids Res. 2017 May 19;45(9):5183-5197. doi: 10.1093/nar/gkx129. Nucleic Acids Res. 2017. PMID: 28334815 Free PMC article.
References
- Alland L, Muhle R, Hou H J, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. - PubMed
- Bannister A J, Kouzarides T. The CBP coactivator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
- Barlev N A, Candau R, Wang L, Darpino P, Silverman N, Berger S L. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. - PubMed
- Berger S L, Pina B, Silverman N, Marcus G A, Agapite J, Reigier J L, Triezenberg S J, Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:251–265. - PubMed
- Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous