A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function - PubMed (original) (raw)
A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function
Z Liu et al. Mol Cell Biol. 1999 Oct.
Abstract
The Hap2,3,4,5p transcription complex is required for expression of many mitochondrial proteins that function in electron transport and the tricarboxylic acid (TCA) cycle. We show that as the cells' respiratory function is reduced or eliminated, the expression of four TCA cycle genes, CIT1, ACO1, IDH1, and IDH2, switches from HAP control to control by three genes, RTG1, RTG2, and RTG3. The expression of four additional TCA cycle genes downstream of IDH1 and IDH2 is independent of the RTG genes. We have previously shown that the RTG genes control the retrograde pathway, defined as a change in the expression of a subset of nuclear genes, e.g., the glyoxylate cycle CIT2 gene, in response to changes in the functional state of mitochondria. We show that the cis-acting sequence controlling RTG-dependent expression of CIT1 includes an R box element, GTCAC, located 70 bp upstream of the Hap2,3,4,5p binding site in the CIT1 upstream activation sequence. The R box is a binding site for Rtg1p-Rtg3p, a heterodimeric, basic helix-loop-helix/leucine zipper transcription factor complex. We propose that in cells with compromised mitochondrial function, the RTG genes take control of the expression of genes leading to the synthesis of alpha-ketoglutarate to ensure that sufficient glutamate is available for biosynthetic processes and that increased flux of the glyoxylate cycle, via elevated CIT2 expression, provides a supply of metabolites entering the TCA cycle sufficient to support anabolic pathways. Glutamate is a potent repressor of RTG-dependent expression of genes encoding both mitochondrial and nonmitochondrial proteins, suggesting that it is a specific feedback regulator of the RTG system.
Figures
FIG. 1
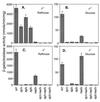
Alternative dependence of CIT1 expression on RTG1, RTG2, RTG3, and HAP2. β-Galactosidase assays were carried out to determine the activity of a CIT1-lacZ reporter gene in wild-type (WT) PSY142 [rho+] and [_rho_0] cells and various mutant derivatives of these strains as indicated. The bp −806 to +9 region of the CIT1 gene was fused to the coding region of the E. coli lacZ gene, and the resulting construct was integrated at the chromosomal CIT1 locus. For each strain grown on either raffinose (YPR) or glucose (YP5%D) medium, four independent transformants were pooled from mid-log-phase cultures and β-galactosidase assays on whole-cell extracts were carried out in triplicate as described in Materials and Methods.
FIG. 2
Glutamate auxotrophy of rtg mutant cells coincides with _RTG_-dependent gene expression. Wild-type (WT) PSY142 [rho+] and [_rho_0] cells and rtg1Δ, rtg2Δ or rtg3Δ mutant derivatives of those strains were streaked on YNBR or YNBD medium with or without 0.02% glutamate. (A) YNBR plus glutamate; (B) YNBR alone; (C) YNBD plus glutamate; (D) YNBD. Only the [_rho_0] mutant derivatives are glutamate auxotrophs in derepressed cells, whereas both [rho+] and [_rho_0] mutant derivatives are glutamate auxotrophs in glucose-repressed cells.
FIG. 3
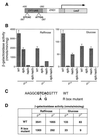
A functional R box in the CIT1 UAS. (A) Diagram of a CIT1 UAS-CYC1-lacZ construct in which a 140-bp fragment of the upstream region of CIT1 from bp −400 to −260 was fused to the transcriptional start site of the CYC1 gene and fused to the reading frame of the E. coli lacZ gene. Positions of the putative Rtg1p-Rtg3p R box binding site, GTCAC, and the Hap2,3,4,5p binding site, ATTGG, are indicated. (B) Wild-type (WT) PSY142 [rho+] and [_rho_0] cells and rtg3Δ, hap2Δ, and rtg3Δ hap2Δ derivatives were transformed with the CIT1 UAS-CYC1-lacZ construct in a centromeric plasmid. Pools of 10 transformants of each were grown to mid-log phase on YNBcasR or YNBcas5%D medium, and β-galactosidase activity was determined in cell-free extracts. (C) Two mutations were introduced into the R box in the CIT1 UAS-CYC1-lacZ construct as indicated in boldface and described in Materials and Methods. This construct was placed into a centromeric plasmid to yield pCIT1(R)(UAS)-CYC1-LacZ. (D) PSY142 [rho+] and [_rho_0] cells were transformed either with pCIT1(UAS)-CYC1-LacZ, containing the wild-type R box, or with pCIT1(R)(UAS)-CYC1-LacZ, containing the mutant R box construct. Ten transformants of each were pooled and grown to mid-log phase in YNBcasR or YNBcas5%D medium, and β-galactosidase activities were measured in cell-free extracts. Standard errors from triplicate assays are <10%.
FIG. 4
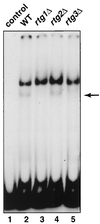
The Rtg1p-Rtg3p complex binds to the CIT1 UAS. Whole-cell extracts were prepared from wild-type (wt) [rho+] PSY142 cells and from rtg1Δ, rtg2Δ, and rtg3Δ derivatives grown on YNB5%D medium supplemented with 0.01% glutamate. EMSA was carried out with a [γ-32P]ATP-labeled 140-bp DNA probe of the CIT1 UAS as described in Materials and Methods. The control lane 1 is the probe alone. The arrow indicates the gel-retarded band whose presence is dependent on Rtg1p and Rtg3p.
FIG. 5
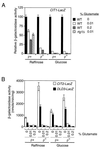
Glutamate is a repressor of _RTG_-dependent gene expression. (A) PSY 142 [rho+] and [_rho_0] strains with an integrated copy of the −806 CIT1-lacZ reporter gene and an rtg1Δ derivative of those strains were grown in YNBR or YNB5%D medium as controls. Parallel cultures contained glutamate in the medium at the indicated concentrations. Whole-cell extracts were prepared, and β-galactosidase activities were determined. Data are expressed as β-galactosidase activities normalized to the value for the wild-type (WT) grown in the absence of glutamate. (B) Same as panel A except that wild-type [rho+] and [_rho_0] strains lacked the CIT1 reporter gene and instead were transformed with centromere-based plasmids containing a CIT2-lacZ reporter gene (18) or a DLD3 reporter gene in which the bp −500 region of DLD3 was fused to lacZ (4).
FIG. 6
Constitutive expression of CIT1 fails to rescue the glutamate auxotrophy and acetate− phenotypes of rtg mutant cells. Wild-type (WT) PSY142 [rho+] cells and cit1Δ, cit2Δ, rtg1Δ, rtg2Δ, or rtg3Δ derivatives were transformed with a centromere-based plasmid, pACT1-CIT1, in which CIT1 expression was placed under the control of the constitutive ACT1 promoter. As controls, these strains were also transformed with pRS416 alone. Cells were streaked on YNBD–0.02% glutamate (A), YNBD (B), YNBcasGly (C) or YNBcasAce (D) medium and grown for 2.5 to 3.5 days at 30°.
FIG. 7
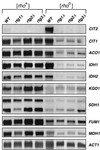
Northern blot analysis of TCA cycle gene expression. Total RNA was prepared from PSY142 [rho+] and [_rho_0] wild-type (WT) strains and their rtg1Δ, rtg2Δ, and rtg3Δ derivatives grown to mid-log phase on YPR medium. Blots were probed for each of the indicated genes as described in Materials and Methods. RNA loads were normalized to the level of transcripts of the ACT1 gene. CIT2 transcripts were also analyzed as a control.
FIG. 8
Model for the role of the RTG genes in the control of expression of TCA and glyoxylate cycle genes. As mitochondrial function decreases (shaded triangle), the expression of CIT1, ACO1, IDH1, and IDH2 becomes increasingly dependent on the RTG genes and less dependent on the HAP genes. Glutamate is shown as a feedback regulator of _RTG_-dependent pathways of gene expression. α-KG, α-ketoglutarate.
Similar articles
- A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus.
Jia Y, Rothermel B, Thornton J, Butow RA. Jia Y, et al. Mol Cell Biol. 1997 Mar;17(3):1110-7. doi: 10.1128/MCB.17.3.1110. Mol Cell Biol. 1997. PMID: 9032238 Free PMC article. - RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus.
Liao X, Butow RA. Liao X, et al. Cell. 1993 Jan 15;72(1):61-71. doi: 10.1016/0092-8674(93)90050-z. Cell. 1993. PMID: 8422683 - Molecular genetics of yeast TCA cycle isozymes.
McAlister-Henn L, Small WC. McAlister-Henn L, et al. Prog Nucleic Acid Res Mol Biol. 1997;57:317-39. doi: 10.1016/s0079-6603(08)60285-8. Prog Nucleic Acid Res Mol Biol. 1997. PMID: 9175438 Review. No abstract available. - SURVEY AND SUMMARY: Saccharomyces cerevisiae basic helix-loop-helix proteins regulate diverse biological processes.
Robinson KA, Lopes JM. Robinson KA, et al. Nucleic Acids Res. 2000 Apr 1;28(7):1499-505. doi: 10.1093/nar/28.7.1499. Nucleic Acids Res. 2000. PMID: 10710415 Free PMC article. Review.
Cited by
- Mathematical modeling and analysis of mitochondrial retrograde signaling dynamics.
Chiu ST, Tseng WW, Wei AC. Chiu ST, et al. iScience. 2022 Nov 11;25(12):105502. doi: 10.1016/j.isci.2022.105502. eCollection 2022 Dec 22. iScience. 2022. PMID: 36444303 Free PMC article. - A novel Rtg2p activity regulates nitrogen catabolism in yeast.
Pierce MM, Maddelein ML, Roberts BT, Wickner RB. Pierce MM, et al. Proc Natl Acad Sci U S A. 2001 Nov 6;98(23):13213-8. doi: 10.1073/pnas.181486098. Epub 2001 Oct 30. Proc Natl Acad Sci U S A. 2001. PMID: 11687605 Free PMC article. - [Gene expression profiling of classic mitochondrial disorders. Its value in finding therapeutic strategies].
Mende S, Storch A, Reichmann H. Mende S, et al. Nervenarzt. 2007 Oct;78(10):1155-9. doi: 10.1007/s00115-007-2266-4. Nervenarzt. 2007. PMID: 17458528 Review. German. - The deletion of the succinate dehydrogenase gene KlSDH1 in Kluyveromyces lactis does not lead to respiratory deficiency.
Saliola M, Bartoccioni PC, De Maria I, Lodi T, Falcone C. Saliola M, et al. Eukaryot Cell. 2004 Jun;3(3):589-97. doi: 10.1128/EC.3.3.589-597.2004. Eukaryot Cell. 2004. PMID: 15189981 Free PMC article. - The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine.
Crespo JL, Powers T, Fowler B, Hall MN. Crespo JL, et al. Proc Natl Acad Sci U S A. 2002 May 14;99(10):6784-9. doi: 10.1073/pnas.102687599. Epub 2002 May 7. Proc Natl Acad Sci U S A. 2002. PMID: 11997479 Free PMC article.
References
- Chelstowska A, Butow R A. RTG genes in yeast that function in communication between mitochondria and the nucleus are also required for expression of genes encoding peroxisomal proteins. J Biol Chem. 1995;270:18141–18146. - PubMed
- Chelstowska, A., Z. Liu, Y. Jia, D. Amberg, and R. A. Butow. Yeast, in press. - PubMed
- Chen D-C, Yang B-C, Kuo T-T. One-step transformation of yeast in stationary phase. Curr Genet. 1992;21:83–84. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous