The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p - PubMed (original) (raw)
The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p
J N McMillan et al. Mol Cell Biol. 1999 Oct.
Abstract
In Saccharomyces cerevisiae, the Wee1 family kinase Swe1p is normally stable during G(1) and S phases but is unstable during G(2) and M phases due to ubiquitination and subsequent degradation. However, perturbations of the actin cytoskeleton lead to a stabilization and accumulation of Swe1p. This response constitutes part of a morphogenesis checkpoint that couples cell cycle progression to proper bud formation, but the basis for the regulation of Swe1p degradation by the morphogenesis checkpoint remains unknown. Previous studies have identified a protein kinase, Hsl1p, and a phylogenetically conserved protein of unknown function, Hsl7p, as putative negative regulators of Swe1p. We report here that Hsl1p and Hsl7p act in concert to target Swe1p for degradation. Both proteins are required for Swe1p degradation during the unperturbed cell cycle, and excess Hsl1p accelerates Swe1p degradation in the G(2)-M phase. Hsl1p accumulates periodically during the cell cycle and promotes the periodic phosphorylation of Hsl7p. Hsl7p can be detected in a complex with Swe1p in cell lysates, and the overexpression of Hsl7p or Hsl1p produces an effective override of the G(2) arrest imposed by the morphogenesis checkpoint. These findings suggest that Hsl1p and Hsl7p interact directly with Swe1p to promote its recognition by the ubiquitination complex, leading ultimately to its destruction.
Figures
FIG. 1
Negative regulation of Swe1p in unperturbed cells by a pathway involving both Hsl1p and Hsl7p. (A) G2 delay (resulting in bud elongation) when SWE1 copy number is doubled in the absence of Hsl1p, Hsl7p, or both. Wild-type (WT) (JMY1469), _hsl1_Δ (JMY1503), _hsl7_Δ (JMY1505), and _hsl1Δ hsl7_Δ (JMY1507) strains and related strains containing an extra copy of SWE1 (JMY1470, JMY1477, JMY1475, and JMY1479) were observed by using differential interference contrast optics during exponential growth (5 × 106 cells/ml) in YEPD medium. (B and C) Genetic interactions among SWE1, HSL1, HSL7, and MIH1. (B) Diploid strains JMY1569 (upper row) and JMY1570 (lower row) were sporulated to generate haploid segregants with the indicated genotypes (confirmed by replica plating and analysis of marker genes). Following tetrad dissection, spores were allowed to grow on YEPD medium for 2 days before the resulting microcolonies were photographed (C). Diploid strains JMY1571 (panels 1 and 2) and JMY1572 (panels 3 and 4) were sporulated, and spores of the indicated genotypes were grown on YEPG medium to induce overexpression of the _GAL_-regulated genes.
FIG. 2
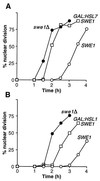
Override of the morphogenesis checkpoint by overexpression of HSL1 or HSL7. (A) Strains DLY657 (cdc24-1 SWE1) (○), DLY690 (_cdc24-1 swe1_Δ) (●), and JMY1284 (cdc24-1 SWE1 GAL:HSL7:LEU2) (□) were grown overnight at 24°C (permissive temperature) in YEPG to induce the GAL promoter, synchronized in G1 phase with α-factor, and released into fresh YEPG at 37°C (restrictive temperature), where actin polarization and bud formation did not occur. At 30-min intervals, cells were fixed and stained to monitor the kinetics of nuclear division; 200 cells were scored in each sample. (B) Strains DLY657 (○), DLY690 (●), and JMY1495 (cdc24-1 SWE1 GAL:HSL1:LEU2) (□) were grown at 24°C in YEPS (noninducing nonrepressing medium for the GAL promoter) and arrested in G1 phase with α-factor. Galactose was then added to induce the GAL promoter, and 1 h later the cells were released into fresh YEPG at 37°C and monitored as described above.
FIG. 3
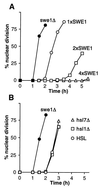
Equivalent checkpoint delays in Hsl+ and Hsl− cells. (A) Strains JMY1472 (cdc24-1 SWE1) (○), DLY690 (_cdc24-1 swe1_Δ) (●), JMY1494 (cdc24-1 2xSWE1) (□), and JMY1493 (cdc24-1 4xSWE1) (▵) were grown at 24°C (permissive temperature), synchronized in G1 phase with α-factor, and released at 37°C (restrictive temperature), where actin polarization and bud formation did not occur. At 30-min intervals, cells were fixed and stained to monitor the kinetics of nuclear division; 200 cells were scored in each sample. (B) Strains DLY657 (cdc24-1 SWE1) (○), DLY690 (●), JMY1300 (_cdc24-1 SWE1 hsl1_Δ) (□), and JMY1301 (_cdc24-1 SWE1 hsl7_Δ) (▵) were synchronized and analyzed as described for panel A.
FIG. 4
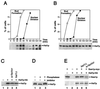
Characterization of Hsl1p and Hsl7p. (A) Periodic accumulation of Hsl1p during the cell cycle. Wild-type cells expressing Hsl1p-myc (strain JMY1500) were grown in YEPD, synchronized in G1 phase with α-factor, and released into a fresh medium. Cells were harvested at the indicated times, and separate aliquots were lysed to detect Hsl1p or fixed to monitor bud formation and nuclear division. Hsl1p-myc was immunoprecipitated from lysates containing 200 μg of total protein, separated by SDS-polyacrylamide gel electrophoresis (PAGE), and immunoblotted with anti-myc antibody. (B to D) Hsl1p-dependent phosphorylation of Hsl7p during the cell cycle. (B) Wild-type cells expressing Hsl7p-HA (strain JMY1521) were synchronized as described above. Lysates containing 20 μg of total protein were separated by SDS-PAGE, and Hsl7p-HA was detected by immunoblotting with anti-HA antibody. (C) Lysates were prepared from an Hsl7p-HA-expressing strain (JMY1521) that had been arrested in G1 phase with α-factor (lane 3) and from cells of strains expressing or lacking Hsl7p-HA, Swe1p-myc (GAL regulated), and Hsl1p as indicated (lane 1, M-1505; lane 2, M-1537; and lane 4, JMY1539) that had been arrested in G2 phase by growth for 3 h after galactose was added to induce overexpression of Swe1p-myc. Proteins were separated by SDS-PAGE and Hsl7p-HA was detected by immunoblotting with anti-HA antibody. (D) Anti-HA immunoprecipitates were prepared from lysate of a strain (M-1537) expressing Hsl7p-HA and divided into three equal aliquots that were subjected to a mock phosphatase treatment (lane 1), treatment with potato acid phosphatase (lane 2), or treatment with phosphatase together with the phosphatase inhibitor sodium orthovanadate (lane 3). Proteins were separated by SDS-PAGE, and Hsl7p-HA was detected by immunoblotting with anti-HA antibody. (E) Coimmunoprecipitation of Hsl7p-HA with Swe1p-myc. Lysates were prepared from strains expressing Hsl7p-HA, Swe1p-myc (GAL regulated), and/or Hsl1p, as indicated (lane 1, M-1505; lane 2, M-1295; lane 3, M-1537; and lane 4, JMY1539), that had been arrested in G2 phase as described for panel C. Lysate was also prepared from a strain expressing both tagged proteins that had been arrested in G1 phase with α-factor (JMY1521 [lane 5]). Anti-myc immunoprecipitates were prepared from samples containing 200 μg of total protein, separated by SDS-PAGE, and immunoblotted with anti-myc (upper blot) or anti-HA (lower blot) antibody.
FIG. 5
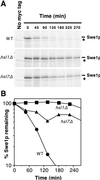
Stabilization of Swe1p in _hsl1_Δ and _hsl7_Δ strains. (A) CDC28Y19F GAL:SWE1myc strains RSY342 (wild type [WT]) (HSL1 HSL7) (top), RSY361 (hsl1Δ HSL7) (middle), and RSY356 (_HSL1 hsl7_Δ) (bottom) were grown in YEPS and induced to express Swe1p-myc by 10 min of growth in the presence of galactose. The cells were harvested, pulse labeled with [35S]methionine and cysteine for 10 min, harvested again, and resuspended in fresh YEPD (to repress the GAL promoter) containing nonradioactive methionine and cysteine. The amounts of 35S-labeled Swe1p-myc were determined at intervals by immunoprecipitation and SDS-PAGE. Cells of a strain (DLY1) not expressing Swe1p-myc were pulse labeled and processed as described above, providing a control shown in the left-hand lane of each gel. The asterisk indicates a labeled band that is present in cells lacking Swe1p-myc (left lanes) and binds to the protein A beads used for immunoprecipitation. (B) The radioactive signals from the gels shown in panel A were quantitated with a phosphorimager. These experiments were performed with CDC28Y19F strains to avoid potential complications arising from the dependence of Swe1p degradation on Cdc28p activity (48); i.e., if the Swe1p produced during the pulse substantially inhibited Cdc28p, an artifactual stabilization of Swe1p might be observed during the chase period. However, Cdc28pY19F, which lacks the Swe1p phosphorylation site, is largely resistant to inhibition by Swe1p. We confirmed that cell proliferation indeed continued through the pulse-chase protocol in all strains (data not shown).
FIG. 6
Acceleration of Swe1p degradation by overexpression of Hsl1p. (A) CDC28Y19F GAL:SWE1myc strains RSY342 (WT), RSY366 (GAL:HSL1), and RSY370 (GAL:HSL7) were grown in YEPS and induced to overexpress the _GAL_-regulated genes by addition of galactose for 3 h. The cells were harvested, pulse labeled with [35S]methionine and cysteine for 10 min, harvested again, and resuspended in fresh YEPG containing nonradioactive methionine and cysteine. The amount of 35S-labeled Swe1p-myc was determined by immunoprecipitation and SDS-PAGE. The asterisk indicates a labeled band that is present in cells lacking Swe1p-myc (left lanes) and binds to the protein A beads used for immunoprecipitation. (B) The radioactive signals from the gels shown in panel A were quantitated with a phosphorimager.
FIG. 7
Cell cycle specificity of the acceleration of Swe1p degradation by overexpression of Hsl1p. CDC28Y19F GAL:SWE1myc strains RSY342 (WT) and RSY366 (GAL:HSL1) were grown in YEPS and induced to overexpress the _GAL_-regulated genes by addition of galactose. Just after galactose addition, the culture was split, and α-factor (50 ng/ml) was added to one set (A) while nocodazole (15 μg/ml) was added to the other (B). After incubation for 4 h, the cells were harvested, pulse labeled with [35S]methionine and cysteine for 10 min, harvested again, and resuspended in fresh YEPG containing nonradioactive methionine and cysteine. The labeling and chase media also contained α-factor or nocodazole to maintain the cell cycle arrest throughout. The amount of 35S-labeled Swe1p-myc remaining was determined by immunoprecipitation and SDS-PAGE.
FIG. 8
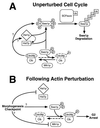
Model for control of the S. cerevisiae cell cycle by the morphogenesis checkpoint. During the unperturbed cell cycle (A), Hsl1p and Hsl7p promote Swe1p hyperphosphorylation (P) leading to recognition by SCFMet30, which catalyzes polyubiquitination (Ub), resulting in the subsequent degradation of Swe1p. Clb-Cdc28p complexes also contribute to Swe1p degradation, acting either through Hsl1p-Hsl7p or separately on Swe1p. Although Hsl7p can bind to Swe1p in the absence of Hsl1p, the fate of the complex may be regulated by Hsl1p-mediated phosphorylation of Hsl7p. The net effect of these interactions is to promote Swe1p degradation in G2/M phase, which promotes the activation of Clb-Cdc28p complexes and hence the unimpeded progression of cells through mitosis. (Note, however, that the activity of Mih1p appears normally to be high enough to keep Clb-Cdc28p largely in the active state even if Swe1p degradation does not occur on schedule.) Following perturbation of the actin cytoskeleton (B), the morphogenesis checkpoint inhibits Hsl1p-Hsl7p, thus preventing Swe1p degradation. However, Swe1p stabilization alone is insufficient to promote G2 arrest, and other checkpoint-responsive pathways must also act to regulate Swe1p and/or Mih1p, so that the balance of their activities is tilted in favor of the phosphorylation and inhibition of Cdc28p, leading to G2 arrest.
Similar articles
- Determinants of Swe1p degradation in Saccharomyces cerevisiae.
McMillan JN, Theesfeld CL, Harrison JC, Bardes ES, Lew DJ. McMillan JN, et al. Mol Biol Cell. 2002 Oct;13(10):3560-75. doi: 10.1091/mbc.e02-05-0283. Mol Biol Cell. 2002. PMID: 12388757 Free PMC article. - Roles of Hsl1p and Hsl7p in Swe1p degradation: beyond septin tethering.
King K, Jin M, Lew D. King K, et al. Eukaryot Cell. 2012 Dec;11(12):1496-502. doi: 10.1128/EC.00196-12. Epub 2012 Oct 5. Eukaryot Cell. 2012. PMID: 23042131 Free PMC article. - Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae.
Longtine MS, Theesfeld CL, McMillan JN, Weaver E, Pringle JR, Lew DJ. Longtine MS, et al. Mol Cell Biol. 2000 Jun;20(11):4049-61. doi: 10.1128/MCB.20.11.4049-4061.2000. Mol Cell Biol. 2000. PMID: 10805747 Free PMC article. - Eavesdropping on the cytoskeleton: progress and controversy in the yeast morphogenesis checkpoint.
Keaton MA, Lew DJ. Keaton MA, et al. Curr Opin Microbiol. 2006 Dec;9(6):540-6. doi: 10.1016/j.mib.2006.10.004. Epub 2006 Oct 19. Curr Opin Microbiol. 2006. PMID: 17055334 Review. - The morphogenesis checkpoint: how yeast cells watch their figures.
Lew DJ. Lew DJ. Curr Opin Cell Biol. 2003 Dec;15(6):648-53. doi: 10.1016/j.ceb.2003.09.001. Curr Opin Cell Biol. 2003. PMID: 14644188 Review.
Cited by
- Identification of C18:1-phytoceramide as the candidate lipid mediator for hydroxyurea resistance in yeast.
Matmati N, Metelli A, Tripathi K, Yan S, Mohanty BK, Hannun YA. Matmati N, et al. J Biol Chem. 2013 Jun 14;288(24):17272-84. doi: 10.1074/jbc.M112.444802. Epub 2013 Apr 25. J Biol Chem. 2013. PMID: 23620586 Free PMC article. - AgSwe1p regulates mitosis in response to morphogenesis and nutrients in multinucleated Ashbya gossypii cells.
Helfer H, Gladfelter AS. Helfer H, et al. Mol Biol Cell. 2006 Oct;17(10):4494-512. doi: 10.1091/mbc.e06-03-0215. Epub 2006 Aug 9. Mol Biol Cell. 2006. PMID: 16899511 Free PMC article. - Determinants of Swe1p degradation in Saccharomyces cerevisiae.
McMillan JN, Theesfeld CL, Harrison JC, Bardes ES, Lew DJ. McMillan JN, et al. Mol Biol Cell. 2002 Oct;13(10):3560-75. doi: 10.1091/mbc.e02-05-0283. Mol Biol Cell. 2002. PMID: 12388757 Free PMC article. - Localization of Saccharomyces cerevisiae protein phosphatase 2A subunits throughout mitotic cell cycle.
Gentry MS, Hallberg RL. Gentry MS, et al. Mol Biol Cell. 2002 Oct;13(10):3477-92. doi: 10.1091/mbc.02-05-0065. Mol Biol Cell. 2002. PMID: 12388751 Free PMC article. - From START to FINISH: the influence of osmotic stress on the cell cycle.
Radmaneshfar E, Kaloriti D, Gustin MC, Gow NA, Brown AJ, Grebogi C, Romano MC, Thiel M. Radmaneshfar E, et al. PLoS One. 2013 Jul 10;8(7):e68067. doi: 10.1371/journal.pone.0068067. Print 2013. PLoS One. 2013. PMID: 23874495 Free PMC article.
References
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM053050/GM/NIGMS NIH HHS/United States
- GM18455/GM/NIGMS NIH HHS/United States
- R37 GM031006/GM/NIGMS NIH HHS/United States
- F32 GM015766/GM/NIGMS NIH HHS/United States
- GM31006/GM/NIGMS NIH HHS/United States
- F32 GM018455/GM/NIGMS NIH HHS/United States
- R01 GM031006/GM/NIGMS NIH HHS/United States
- GM15766/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases