Heteromeric kainate receptors formed by the coassembly of GluR5, GluR6, and GluR7 - PubMed (original) (raw)
Heteromeric kainate receptors formed by the coassembly of GluR5, GluR6, and GluR7
C Cui et al. J Neurosci. 1999.
Abstract
In the CNS kainate subtype glutamate receptors (GluRs) are likely to be heteromeric assemblies containing multiple gene products. However, although recombinant kainate receptors from the GluR5-GluR7 gene family have been studied extensively in their homomeric forms, there have been no tests to determine whether these subunits can coassemble with each other. We used the GluR5 selective agonists (RS)-2-amino-3-(3-hydroxy-5-tertbutylisoxazol-4-yl)propanoic acid (ATPA) and (S)-5-iodowillardiine (I-will) to test for the coassembly of GluR5 with GluR6 and GluR7 by measuring changes in rectification that occur for heteromeric receptors containing both edited and unedited Q/R site subunits. Birectifying ATPA and I-will responses resulting from polyamine block for homomeric GluR5(Q) became outwardly rectifying when GluR6(R) was coexpressed with GluR5(Q), although GluR6 was not activated by ATPA or I-will, indicating the formation of heteromeric receptors. Similar approaches showed the coassembly of GluR7 with GluR6 and GluR5. Heteromeric kainate receptors containing both GluR5 and GluR6 subunits exhibited novel functional properties, including reduced desensitization and faster recovery from desensitization than those recorded for homomeric GluR5. Coexpression of GluR6 with GluR5 also enhanced the magnitude of responses to GluR5 selective agonists. In contrast, the coassembly of GluR7 with GluR6 markedly decreased the amplitude of agonist responses. Our results indicate that, similar to AMPA receptors, the kainate receptor subunits GluR5-GluR7 exhibit promiscuous coassembly. The formation of heteromeric kainate receptors may help to explain why the functional properties of native kainate receptors differ from those that have been reported for recombinant kainate receptors.
Figures
Fig. 1.
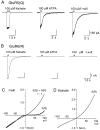
Selective activation of GluR5 by ATPA and I-will.A, Responses from a HEK cell transfected with GluR5(Q) to 100 μ
m
kainate, 10 μ
m
ATPA, and 100 μ
m
I-will at −60 mV. B, When the same agonists were applied to a cell transfected with GluR6(R), only kainate produced inward current responses. C, Ramp_I–V_ plots for responses to 100 μ
m
I-will recorded from HEK cells transfected with GluR5(Q) or both GluR5(Q) and GluR6(R) at a cDNA ratio of 1:1. D, Ramp_I–V_ plot for responses to 100 μ
m
kainate recorded from a HEK cell transfected with GluR6(R). In all experiments the cells were treated with concanavalin A to attenuate desensitization.
Fig. 2.
Functional overexpression of GluR6 dominates heteromer formation with GluR5. Shown are G–V plots for responses to 10 μ
m
ATPA (A) and 100 μ
m
I-will (B, C) recorded from HEK cells transfected with homomeric GluR5(Q) or GluR5(Q) plus GluR6(R) at cDNA ratios of 1:1, 2:1, 5:1, and 20:1, as indicated. In _A_and B, the data points show the mean ± SEM for 4–12 cells per experiment; at cDNA ratios for GluR5(Q) to GluR6(R) of 1:1, 2:1, and 5:1 the responses to GluR5 selective agonists show a similar weak outward rectification. C,G–V plots for individual cells normalized to the conductance at −100 mV; at cDNA ratios of 20:1 the rectification varied from cell to cell.
Fig. 3.
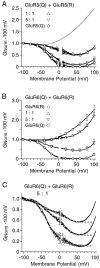
Coassembly of edited and unedited GluR5 or GluR6 generates multiple families of kainate receptors.A, G–V plots of the responses to 100 μ
m
kainate for homomeric GluR5(Q) or GluR5(Q) plus GluR5(R) transfected at cDNA ratios of 1:1 and 5:1. B,G–V plots for homomeric GluR6(Q), homomeric GluR6(R), or GluR6(Q) plus GluR6(R) transfected at cDNA ratios of 1:1 and 5:1. The symbols show the mean ± SEM of responses to 100 μ
m
kainate for 5–11 cells per experiment; the_dotted line_ in A shows the responses for homomeric GluR6(R). C, Responses to 50 μ
m
domoate for three cells transfected with GluR6(Q) plus GluR6(R) at a cDNA ratio of 5:1; open circles indicate the fits of the sum of two Boltzmann functions over the range of −100 to +50 mV.
Fig. 4.
Coassembly of GluR5 and GluR6 upregulates the responses to GluR5 selective agonists. A,G–V plots for responses to 100 μ
m
I-will and 100 μ
m
kainate scaled to have the same amplitude at −100 mV for cells transfected with both GluR5(R) with GluR6(Q) at a cDNA ratio of 1:1; the symbols show the mean ± SEM of responses for six cells; the unscaled response to I-will is plotted as a dotted line. B, _G–V_plots for responses to 100 μ
m
I-will and 100 μ
m
kainate normalized to have the same amplitude at −100 mV for cells transfected with both GluR5(Q) and GluR6(R) at a cDNA ratio of 1:1. The symbols show the mean ± SEM of responses for nine cells; the unscaled response to I-will is plotted as a dotted line. C, Box plots of the mean amplitude of I-will-evoked currents at −100 mV for the edited and unedited versions of GluR5 expressed alone or with GluR6 at a cDNA ratio of 1:1, as indicated; the cells were treated with concanavalin A to attenuate desensitization. The top and bottom boundaries of the boxes indicate 25 and 75% of the data, and the whiskers indicate 10 and 90% of the data. The median is indicated by a bold bar; the_shaded areas_ indicate the mean ± SD. Note that the coexpression of GluR6 upregulates the amplitude of responses to I-will.
Fig. 5.
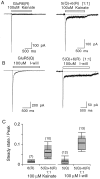
Coassembly of GluR5 and GluR6 reduces kainate receptor desensitization. A, Responses to 100 μ
m
kainate recorded from HEK cells expressing homomeric GluR6(R) or GluR5(Q) plus GluR6(R) at a cDNA ratio of 1:1.B, Responses to 100 μ
m
I-will recorded from HEK cells expressing homomeric GluR5(Q) or GluR5(Q) plus GluR6(R) at a cDNA ratio of 1:1. C, Desensitization, expressed as a steady-state/peak current, for homomeric GluR6(R), homomeric GluR5(Q), and GluR5(Q) plus GluR6(R) heteromers. The steady-state current amplitude was measured at 1 sec after the start of the application of the agonists. The box plots were generated by the method described in Figure 4_C_.
Fig. 6.
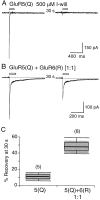
Coassembly of GluR5 and GluR6 speeds recovery from desensitization. A, Responses to paired applications of 500 μ
m
I-will separated by 30 sec intervals reveal persistent desensitization for homomeric GluR5(Q). B, Recovery from desensitization is much faster when GluR5(Q) is coexpressed with GluR6(R) at a 1:1 ratio of cDNAs. _C,Box plots for recovery from desensitization measured at 30 sec intervals between paired applications of I-will. The box plots were generated by the method described in Figure 4_C.
Fig. 7.
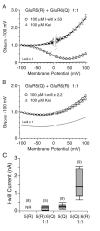
Coassembly of GluR5 and GluR6 with GluR7.A, G–V plots of responses to 100 μ
m
I-will for homomeric GluR5(Q) or GluR5(Q) and GluR7(R) transfected at cDNA ratios of 1:1 and 5:1; the _symbols_indicate the mean ± SEM for three to five cells per experiment.B, Mean amplitude of responses to 100 μ
m
kainate and 100 μ
m
I-will recorded from cells transfected with homomeric GluR5(Q) or GluR5(Q) plus GluR7(R) at cDNA ratios of 5:1 and 1:1; the error bars indicate the mean ± SEM.C, G–V plots of responses to 500 μ
m
kainate for homomeric GluR6(Q590E) or GluR7(R) and GluR6(Q590E) transfected at a cDNA ratio of 1:1; the_symbols_ indicate the mean ± SEM for five to six cells per experiment. D, Mean amplitude of responses to 500 μ
m
kainate recorded from cells transfected with homomeric GluR6(Q590E) or GluR7(R) plus GluR6(Q590E); the error bars indicate the mean ± SEM. In all experiments the cells were treated with concanavalin A to attenuate desensitization.
Fig. 8.
Molecular mechanisms for kainate receptor diversity. The scheme illustrates possible receptor combinations that could be formed by coassembly of the top row, GluR5(Q) with GluR6(R), and the bottom row, GluR5(R) with GluR6(Q), assuming a tetrameric stoichiometry._I_max indicates the maximum response that could be generated by a GluR5 selective agonist according to the values published by Rosenmund et al. (1998) for the activation of homomeric AMPA receptors at the appropriate receptor occupancies by GluR5 selective agonists; NR indicates that these forms would not be expected to respond to I-will or ATPA. Mean γ indicates the maximum values for single-channel conductance recorded bySwanson et al. (1996) for the Q and R forms of homomeric GluR5 and GluR6.
Similar articles
- GluR5 and GluR6 kainate receptor subunits coexist in hippocampal neurons and coassemble to form functional receptors.
Paternain AV, Herrera MT, Nieto MA, Lerma J. Paternain AV, et al. J Neurosci. 2000 Jan 1;20(1):196-205. doi: 10.1523/JNEUROSCI.20-01-00196.2000. J Neurosci. 2000. PMID: 10627597 Free PMC article. - Pharmacological characterization of glutamatergic agonists and antagonists at recombinant human homomeric and heteromeric kainate receptors in vitro.
Alt A, Weiss B, Ogden AM, Knauss JL, Oler J, Ho K, Large TH, Bleakman D. Alt A, et al. Neuropharmacology. 2004 May;46(6):793-806. doi: 10.1016/j.neuropharm.2003.11.026. Neuropharmacology. 2004. PMID: 15033339 - Effect of RNA editing and subunit co-assembly single-channel properties of recombinant kainate receptors.
Swanson GT, Feldmeyer D, Kaneda M, Cull-Candy SG. Swanson GT, et al. J Physiol. 1996 Apr 1;492 ( Pt 1)(Pt 1):129-42. doi: 10.1113/jphysiol.1996.sp021295. J Physiol. 1996. PMID: 8730589 Free PMC article. - New developments in the molecular pharmacology of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate and kainate receptors.
Fletcher EJ, Lodge D. Fletcher EJ, et al. Pharmacol Ther. 1996;70(1):65-89. doi: 10.1016/0163-7258(96)00014-9. Pharmacol Ther. 1996. PMID: 8804111 Review. - Kainate receptors: pharmacology, function and therapeutic potential.
Jane DE, Lodge D, Collingridge GL. Jane DE, et al. Neuropharmacology. 2009 Jan;56(1):90-113. doi: 10.1016/j.neuropharm.2008.08.023. Epub 2008 Aug 28. Neuropharmacology. 2009. PMID: 18793656 Review.
Cited by
- GluR7 is an essential subunit of presynaptic kainate autoreceptors at hippocampal mossy fiber synapses.
Pinheiro PS, Perrais D, Coussen F, Barhanin J, Bettler B, Mann JR, Malva JO, Heinemann SF, Mulle C. Pinheiro PS, et al. Proc Natl Acad Sci U S A. 2007 Jul 17;104(29):12181-6. doi: 10.1073/pnas.0608891104. Epub 2007 Jul 9. Proc Natl Acad Sci U S A. 2007. PMID: 17620617 Free PMC article. - Presynaptic kainate receptors regulate spinal sensory transmission.
Kerchner GA, Wilding TJ, Li P, Zhuo M, Huettner JE. Kerchner GA, et al. J Neurosci. 2001 Jan 1;21(1):59-66. doi: 10.1523/JNEUROSCI.21-01-00059.2001. J Neurosci. 2001. PMID: 11150320 Free PMC article. - Functional diversity and developmental changes in rat neuronal kainate receptors.
Wilding TJ, Huettner JE. Wilding TJ, et al. J Physiol. 2001 Apr 15;532(Pt 2):411-21. doi: 10.1111/j.1469-7793.2001.0411f.x. J Physiol. 2001. PMID: 11306660 Free PMC article. - Role of GluK1 kainate receptors in seizures, epileptic discharges, and epileptogenesis.
Fritsch B, Reis J, Gasior M, Kaminski RM, Rogawski MA. Fritsch B, et al. J Neurosci. 2014 Apr 23;34(17):5765-75. doi: 10.1523/JNEUROSCI.5307-13.2014. J Neurosci. 2014. PMID: 24760837 Free PMC article. - The role of GLU K5-containing kainate receptors in entorhinal cortex gamma frequency oscillations.
Stanger HL, Alford R, Jane DE, Cunningham MO. Stanger HL, et al. Neural Plast. 2008;2008:401645. doi: 10.1155/2008/401645. Epub 2008 Nov 17. Neural Plast. 2008. PMID: 19043593 Free PMC article.
References
- Belcher SM, Howe JR. Characterization of RNA editing of the glutamate-receptor subunits GluR5 and GluR6 in granule cells during cerebellar development. Brain Res Mol Brain Res. 1997;52:130–138. - PubMed
- Bettler B, Boulter J, Hermans-Borgmeyer I, O’Shea-Greenfield A, Deneris ES, Moll C, Borgmeyer U, Hollmann M, Heinemann S. Cloning of a novel glutamate receptor subunit, GluR5: expression in the nervous system during development. Neuron. 1990;5:583–595. - PubMed
- Bischoff S, Barhanin J, Bettler B, Mulle C, Heinemann S. Spatial distribution of kainate receptor subunit mRNA in the mouse basal ganglia and ventral mesencephalon. J Comp Neurol. 1997;379:541–562. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources