Two family B DNA polymerases from Aeropyrum pernix, an aerobic hyperthermophilic crenarchaeote - PubMed (original) (raw)
Two family B DNA polymerases from Aeropyrum pernix, an aerobic hyperthermophilic crenarchaeote
I K Cann et al. J Bacteriol. 1999 Oct.
Abstract
DNA polymerase activities in fractionated cell extract of Aeropyrum pernix, a hyperthermophilic crenarchaeote, were investigated. Aphidicolin-sensitive (fraction I) and aphidicolin-resistant (fraction II) activities were detected. The activity in fraction I was more heat stable than that in fraction II. Two different genes (polA and polB) encoding family B DNA polymerases were cloned from the organism by PCR using degenerated primers based on the two conserved motifs (motif A and B). The deduced amino acid sequences from their entire coding regions contained all of the motifs identified in family B DNA polymerases for 3'-->5' exonuclease and polymerase activities. The product of polA gene (Pol I) was aphidicolin resistant and heat stable up to 80 degrees C. In contrast, the product of polB gene (Pol II) was aphidicolin sensitive and stable at 95 degrees C. These properties of Pol I and Pol II are similar to those of fractions II and I, respectively, and moreover, those of Pol I and Pol II of Pyrodictium occultum. The deduced amino acid sequence of A. pernix Pol I exhibited the highest identities to archaeal family B DNA polymerase homologs found only in the crenarchaeotes (group I), while Pol II exhibited identities to homologs found in both euryarchaeotes and crenarchaeotes (group II). These results provide further evidence that the subdomain Crenarchaeota has two family B DNA polymerases. Furthermore, at least two DNA polymerases work in the crenarchaeal cells, as found in euryarchaeotes, which contain one family B DNA polymerase and one heterodimeric DNA polymerase of a novel family.
Figures
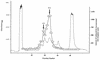
FIG. 1
Chromatographic profile of A. pernix cell extract determined on an anion-exchange column (HiTrap Q) and corresponding DNA polymerase activity. Procedures are described in Materials and Methods —, absorbance at 280 nm monitored throughout the chromatography; □, incorporation of [3H]dTTP into activated DNA.
FIG. 2
Effects of aphidicolin and preincubation temperature on DNA polymerizing activity. The standard assay mixture including fraction I (F1) or II (F2) was incubated at 70°C for 5 min in the presence of the indicated levels of aphidicolin (a), or the enzyme fractions were incubated at the indicated temperatures for 30 min before the activities were measured under the conditions described above (b). P. furiosus (Pfu) Pol I served as a control.
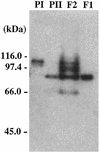
FIG. 3
Activity gel analysis of DNA polymerizing activities in fractionated A. pernix cell extracts, fraction I and fraction II, and recombinant Pol I and Pol II. DNA polymerizing activity was detected by in situ incorporation of [α-32P]dCTP as described in Materials and Methods. Proteins in fraction I (F1), fraction II (F2), purified Pol I (PI), and Pol II (PII) were separated by SDS-PAGE, and 32P incorporated into DNA strands in the gel was detected by autoradiography. The molecular masses indicated on the left were derived from a size standard marker (Bio-Rad, Hercules, Calif.).
FIG. 4
Nucleotide and deduced amino acid sequences of A. pernix polA (a) and polB (b) genes. The regions used in designing primers for genome walking PCR are indicated by black arrows; grey arrows indicated the primers used in amplifying the structural gene for each polymerase. The regions including the exonuclease motifs (3) and the polymerase motifs (32) are underlined by solid and broken lines, respectively; boldface letters represent putative box A sequences.
FIG. 4
Nucleotide and deduced amino acid sequences of A. pernix polA (a) and polB (b) genes. The regions used in designing primers for genome walking PCR are indicated by black arrows; grey arrows indicated the primers used in amplifying the structural gene for each polymerase. The regions including the exonuclease motifs (3) and the polymerase motifs (32) are underlined by solid and broken lines, respectively; boldface letters represent putative box A sequences.
FIG. 5
Production of A. pernix Pol I and Pol II. Proteins were loaded on an SDS–10% polyacrylamide gel and stained with Coomassie brilliant blue. Lanes: 1, molecular mass markers (sizes are indicated on the left); 2, Pol I; 3, Pol II.
FIG. 6
Effects of aphidicolin and preincubation temperature on DNA polymerizing activity of A. pernix Pol I and Pol II. Aliquots of purified A. pernix (Ape) Pol I or Pol II were incubated at 70°C for 5 min in the presence of the indicated levels of aphidicolin (a), or the enzyme fractions were incubated at indicated temperatures for 30 min before the activities were measured under the conditions described above (b). P. furiosus (Pfu) Pol I served as a control.
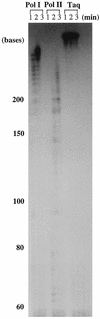
FIG. 7
Chain elongation ability of A. pernix Pol I and Pol II. A 32P-labeled primer annealed to M13 DNA served as the template. Aliquots (5 μl) of reaction mixture were sampled at 1, 2, and 3 min after initiation of reaction, and their products were resolved on an 8% polyacrylamide gel containing 8 M urea. Taq DNA polymerase was used as a control.
Similar articles
- The hyperthermophilic archaeon Pyrodictium occultum has two alpha-like DNA polymerases.
Uemori T, Ishino Y, Doi H, Kato I. Uemori T, et al. J Bacteriol. 1995 Apr;177(8):2164-77. doi: 10.1128/jb.177.8.2164-2177.1995. J Bacteriol. 1995. PMID: 7721707 Free PMC article. - Three proliferating cell nuclear antigen-like proteins found in the hyperthermophilic archaeon Aeropyrum pernix: interactions with the two DNA polymerases.
Daimon K, Kawarabayasi Y, Kikuchi H, Sako Y, Ishino Y. Daimon K, et al. J Bacteriol. 2002 Feb;184(3):687-94. doi: 10.1128/JB.184.3.687-694.2002. J Bacteriol. 2002. PMID: 11790738 Free PMC article. - Cloning and characterization of a family B DNA polymerase from the hyperthermophilic crenarchaeon Pyrobaculum islandicum.
Kähler M, Antranikian G. Kähler M, et al. J Bacteriol. 2000 Feb;182(3):655-63. doi: 10.1128/JB.182.3.655-663.2000. J Bacteriol. 2000. PMID: 10633098 Free PMC article. - The euryarchaeotes, a subdomain of Archaea, survive on a single DNA polymerase: fact or farce?
Ishino Y, Cann IK. Ishino Y, et al. Genes Genet Syst. 1998 Dec;73(6):323-36. doi: 10.1266/ggs.73.323. Genes Genet Syst. 1998. PMID: 10333564 Review. - Archaeal DNA replication: identifying the pieces to solve a puzzle.
Cann IK, Ishino Y. Cann IK, et al. Genetics. 1999 Aug;152(4):1249-67. doi: 10.1093/genetics/152.4.1249. Genetics. 1999. PMID: 10430556 Free PMC article. Review.
Cited by
- A transposon-derived DNA polymerase from Entamoeba histolytica displays intrinsic strand displacement, processivity and lesion bypass.
Pastor-Palacios G, López-Ramírez V, Cardona-Felix CS, Brieba LG. Pastor-Palacios G, et al. PLoS One. 2012;7(11):e49964. doi: 10.1371/journal.pone.0049964. Epub 2012 Nov 30. PLoS One. 2012. PMID: 23226232 Free PMC article. - A korarchaeal genome reveals insights into the evolution of the Archaea.
Elkins JG, Podar M, Graham DE, Makarova KS, Wolf Y, Randau L, Hedlund BP, Brochier-Armanet C, Kunin V, Anderson I, Lapidus A, Goltsman E, Barry K, Koonin EV, Hugenholtz P, Kyrpides N, Wanner G, Richardson P, Keller M, Stetter KO. Elkins JG, et al. Proc Natl Acad Sci U S A. 2008 Jun 10;105(23):8102-7. doi: 10.1073/pnas.0801980105. Epub 2008 Jun 5. Proc Natl Acad Sci U S A. 2008. PMID: 18535141 Free PMC article. - DNA polymerases as useful reagents for biotechnology - the history of developmental research in the field.
Ishino S, Ishino Y. Ishino S, et al. Front Microbiol. 2014 Aug 29;5:465. doi: 10.3389/fmicb.2014.00465. eCollection 2014. Front Microbiol. 2014. PMID: 25221550 Free PMC article. Review. - Unique substrate spectrum and PCR application of Nanoarchaeum equitans family B DNA polymerase.
Choi JJ, Song JG, Nam KH, Lee JI, Bae H, Kim GA, Sun Y, Kwon ST. Choi JJ, et al. Appl Environ Microbiol. 2008 Nov;74(21):6563-9. doi: 10.1128/AEM.00624-08. Epub 2008 Sep 12. Appl Environ Microbiol. 2008. PMID: 18791030 Free PMC article. - Functional interactions of a homolog of proliferating cell nuclear antigen with DNA polymerases in Archaea.
Cann IK, Ishino S, Hayashi I, Komori K, Toh H, Morikawa K, Ishino Y. Cann IK, et al. J Bacteriol. 1999 Nov;181(21):6591-9. doi: 10.1128/JB.181.21.6591-6599.1999. J Bacteriol. 1999. PMID: 10542158 Free PMC article.
References
- Bernad A, Blanco L, Lazaro J M, Martin G, Salas M. A conserved 3′→5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. - PubMed
- Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Presley E A, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Hurst M A, Roberts K M, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. - PubMed
- Cann, I. K. O., S. Ishino, I. Hayashi, K. Komori, H. Toh, K. Morikawa, and Y. Ishino. Unpublished data.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources