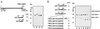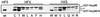Autoproteolysis in nucleoporin biogenesis - PubMed (original) (raw)
Autoproteolysis in nucleoporin biogenesis
J S Rosenblum et al. Proc Natl Acad Sci U S A. 1999.
Abstract
We have molecularly characterized a proteolytic cleavage in conserved nuclear pore complex proteins. This cleavage, previously demonstrated to be essential for the biogenesis of two nuclear pore complex proteins in mammals (Nup98 and Nup96) and yeast (Nup145-N and Nup145-C), occurs between Phe and Ser residues within a highly conserved domain in a polyprotein precursor. Here, we show that a protease is not involved in the cleavage event. By using a combination of domain mapping and site-directed mutagenesis, we demonstrate that the human nuclear pore complex protein Nup98 specifically cleaves itself between F863 and S864. A region of Nup98, amino acids 715-920, is able to cleave, whereas a smaller region, amino acids 772-920, does not cleave. In addition, we have generated a Nup98 mutant that cleaves under defined conditions in vitro. Further, the two cleaved fragments of Nup98 form a complex, providing a possible mechanism whereby specific, yet low-affinity, binding between Nup98 and Nup96 is responsible for the nuclear targeting of Nup96. Although apparently unrelated evolutionarily, Nup98 has converged on an autoproteolytic biogenesis mechanism similar to that of hedgehog proteins, the inteins, and the N-terminal nucleophile proteins.
Figures
Figure 1
A minimal domain for Nup98 cleavage. (A) Full-length Nup98 and N-terminal deletions are represented at left, the cleavage site is indicated (↓), as are the N and C termini. The deletions were made in either wild-type Nup98 or a mutant Nup98 that is unable to cleave. Purified proteins were separated by SDS/12.5% PAGE and visualized with Coomassie blue R-250. The wild-type and mutant constructs of Nup98 (715–920) are in lanes 1 and 2, and the wild-type and mutant constructs of Nup98 (772–920) are in lanes 3 and 4. (B) N-terminal deletions (lanes 5–7) and site-directed mutants (lanes 8–11) are represented at left. The cleavage site and termini are represented, and the region of the C terminus that was subjected to mutagenesis is expanded. Mutagenized amino acids are underlined. Proteins were purified from similar culture volumes, separated by SDS/12.5% PAGE, and visualized with Coomassie blue R-250. GST, glutathione _S_-transferase; GFP, green fluorescent protein.
Figure 2
The minimal cleavage domain of human Nup98 is evolutionarily conserved. The corresponding regions of the human (hs), rat (rn), Caenorhabditis elegans, A. thaliana, and S. cerevisiae proteins are aligned (GenBank accession numbers U41815, P49793, AAA91249, AAD32891, and P49687). Residues common to at least three of the organisms are highlighted. The cleavage site and HFS tripeptide are indicated.
Figure 3
Mutations in the amino acids immediately preceding and following the Nup98 cleavage site. The wild-type protein is in the first lane, followed by seven mutants in amino acid 864 and seven mutants in amino acid 863. The cleavage-site tripeptide is above the lanes, where X is the amino acid directly below the lane. A whole cell extract was prepared, and proteins were separated by SDS/10% PAGE. GST-containing fusion proteins were visualized by immunoblotting. The band between the uncleaved and cleaved proteins (∗) most likely corresponds to a faster migrating conformation of the uncleaved protein.
Figure 4
Cleavage of H862Q-containing mutants. Purified Nup98(H862Q) and Nup98(H862Q/S864C) are in lanes 1 and 2. Nup98(H862Q/S864C) was incubated at ambient temperature for 20 hr with 25, 50, and 200 mM DTT (lanes 3, 4, and 5, respectively). SDS-denatured and native Nup98(H862Q/S864C) were incubated with 200 mM NH2OH at room temperature for 15 min (lanes 6 and 7) and 15 hr (lanes 8 and 9). Additionally, the contrast of the lower region of lanes 3–9 was enhanced (Lower) to facilitate detection of Nup98-C. Proteins were separated by SDS/4–12% NuPAGE (NOVEX, San Diego) and visualized with Coomassie blue R-250.
Figure 5
Proteins capable of _cis_-autoproteolysis. One protein from each autoproteolytic subgroup is schematically represented. Cleavage sites (↓) and adjacent amino acids are indicated.
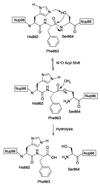
Figure 6
The proposed mechanism for the autoproteolysis of Nup98 and Nup98–Nup96. (The tetrahedral hydroxyoxazolidine intermediate between the precursor and the ester has been omitted for clarity.)
Similar articles
- A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96.
Fontoura BM, Blobel G, Matunis MJ. Fontoura BM, et al. J Cell Biol. 1999 Mar 22;144(6):1097-112. doi: 10.1083/jcb.144.6.1097. J Cell Biol. 1999. PMID: 10087256 Free PMC article. - Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes.
Griffis ER, Xu S, Powers MA. Griffis ER, et al. Mol Biol Cell. 2003 Feb;14(2):600-10. doi: 10.1091/mbc.e02-09-0582. Mol Biol Cell. 2003. PMID: 12589057 Free PMC article. - Molecular determinants of binding between Gly-Leu-Phe-Gly nucleoporins and the nuclear pore complex.
Ratner GA, Hodel AE, Powers MA. Ratner GA, et al. J Biol Chem. 2007 Nov 23;282(47):33968-76. doi: 10.1074/jbc.M707911200. Epub 2007 Sep 26. J Biol Chem. 2007. PMID: 17897945 - Nucleoporin Nup98: a gatekeeper in the eukaryotic kingdoms.
Iwamoto M, Asakawa H, Hiraoka Y, Haraguchi T. Iwamoto M, et al. Genes Cells. 2010 Jun;15(7):661-9. doi: 10.1111/j.1365-2443.2010.01415.x. Epub 2010 Jun 7. Genes Cells. 2010. PMID: 20545767 Review. - The nuclear pore complex.
Heese-Peck A, Raikhel NV. Heese-Peck A, et al. Plant Mol Biol. 1998 Sep;38(1-2):145-62. Plant Mol Biol. 1998. PMID: 9738965 Review.
Cited by
- "Off-pore" nucleoporins relocalize heterochromatic breaks through phase separation.
Merigliano C, Ryu T, See CD, Caridi CP, Li X, Butova NL, Reynolds TW, Deng C, Chenoweth DM, Capelson M, Chiolo I. Merigliano C, et al. bioRxiv [Preprint]. 2024 Jul 18:2023.12.07.570729. doi: 10.1101/2023.12.07.570729. bioRxiv. 2024. PMID: 39071440 Free PMC article. Preprint. - Nuclear transport is becoming crystal clear.
Madrid AS, Weis K. Madrid AS, et al. Chromosoma. 2006 Apr;115(2):98-109. doi: 10.1007/s00412-005-0043-3. Epub 2006 Jan 19. Chromosoma. 2006. PMID: 16421734 Review. No abstract available. - Structural constraints on autoprocessing of the human nucleoporin Nup98.
Sun Y, Guo HC. Sun Y, et al. Protein Sci. 2008 Mar;17(3):494-505. doi: 10.1110/ps.073311808. Protein Sci. 2008. PMID: 18287282 Free PMC article. - Exploring the role of conformational heterogeneity in cis-autoproteolytic activation of ThnT.
Buller AR, Freeman MF, Schildbach JF, Townsend CA. Buller AR, et al. Biochemistry. 2014 Jul 8;53(26):4273-81. doi: 10.1021/bi500385d. Epub 2014 Jun 26. Biochemistry. 2014. PMID: 24933323 Free PMC article. - Molecular basis for the anchoring of proto-oncoprotein Nup98 to the cytoplasmic face of the nuclear pore complex.
Stuwe T, von Borzyskowski LS, Davenport AM, Hoelz A. Stuwe T, et al. J Mol Biol. 2012 Jun 22;419(5):330-46. doi: 10.1016/j.jmb.2012.03.024. Epub 2012 Apr 2. J Mol Biol. 2012. PMID: 22480613 Free PMC article.
References
- Mattaj I W, Englmeier L. Annu Rev Biochem. 1998;67:265–306. - PubMed
- Pemberton L F, Blobel G, Rosenblum J S. Curr Opin Cell Biol. 1998;10:392–399. - PubMed
- Wozniak R W, Rout M P, Aitchison J D. Trends Cell Biol. 1998;8:184–188. - PubMed
- Adam S A. Curr Opin Cell Biol. 1999;11:402–406. - PubMed
- Moore M S. J Biol Chem. 1998;273:22857–22860. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases