Thermodynamics of T cell receptor binding to peptide-MHC: evidence for a general mechanism of molecular scanning - PubMed (original) (raw)
Thermodynamics of T cell receptor binding to peptide-MHC: evidence for a general mechanism of molecular scanning
J J Boniface et al. Proc Natl Acad Sci U S A. 1999.
Abstract
Antigen-dependent activation of T lymphocytes requires T cell receptor (TCR)-mediated recognition of specific peptides, together with the MHC molecules to which they are bound. To achieve this recognition in a reasonable time frame, the TCR must scan and discriminate rapidly between thousands of MHC molecules differing from each other only in their bound peptides. Kinetic analysis of the interaction between a TCR and its cognate peptide-MHC complex indicates that both association and dissociation depend heavily on the temperature, indicating the presence of large energy barriers in both phases. Thermodynamic analysis reveals changes in heat capacity and entropy that are characteristic of protein-ligand associations in which local folding is coupled to binding. Such an "induced-fit" mechanism is characteristic of sequence-specific DNA-binding proteins that must also recognize specific ligands in the presence of a high background of competing elements. Here, we propose that induced fit may endow the TCR with its requisite discriminatory capacity and suggest a model whereby the loosely structured antigen-binding loops of the TCR rapidly explore peptide-MHC complexes on the cell surface until some critical structural complementarity is achieved through localized folding transitions. We further suggest that conformational changes, implicit in this model, may also propagate beyond the TCR antigen-binding site and directly affect self-association of ligated TCRs or TCR-CD3 interactions required for signaling.
Figures
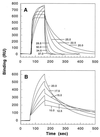
Figure 1
Binding of 2B4 to MCC–Ek monitored at different temperatures on a BIAcore device. Injection of TCR (0.45 mg/ml) was from ≈70 to ≈160 sec, after which dissociation was initiated. Each curve in A (20–37°C) can be distinguished by the faster approach to equilibrium at higher temperatures. Binding curves in A are not directly comparable to those in B (10–20°C), because they were obtained in separate experiments, on different instruments, and at slightly different densities of immobilized ligand. RU, resonance units.
Figure 2
Temperature dependence of 2B4–MCC–Ek association-rate (A) and dissociation-rate (B) constants. (Inset) Variation of the reaction half-life with temperature. Data points shown represent the mean value of three to seven independent measurements with at least three different analyte concentrations. Error bars represent SEM; uncertainty involved in temperature setting was ±0.1°C. Data were fit to a rearranged form of Eq. 1 (detailed in Materials and Methods) with a reference temperature _T_0 of 298.15 K. Note, we chose to illustrate the data by using temperature on the x axis, as opposed to the reciprocal of temperature that is common to Van’t Hoff analysis.
Figure 3
The thermodynamics of the 2B4–MCC–Ek association. (A) Δ_G_°assoc data were fit to Eq. 1 (T_0 = 298.15 K) to yield the parameters and the best-fit line shown. Errors for the indicated parameters represent the estimated SEM obtained from data fitting. (B) Δ_H° and T_Δ_S° values were derived from Eqs. 2 and 3 detailed in Materials and Methods.
Similar articles
- Geometrical characterization of T cell receptor binding modes reveals class-specific binding to maximize access to antigen.
Singh NK, Abualrous ET, Ayres CM, Noé F, Gowthaman R, Pierce BG, Baker BM. Singh NK, et al. Proteins. 2020 Mar;88(3):503-513. doi: 10.1002/prot.25829. Epub 2019 Oct 21. Proteins. 2020. PMID: 31589793 Free PMC article. - T cell receptor interaction with peptide/major histocompatibility complex (MHC) and superantigen/MHC ligands is dominated by antigen.
Ehrich EW, Devaux B, Rock EP, Jorgensen JL, Davis MM, Chien YH. Ehrich EW, et al. J Exp Med. 1993 Aug 1;178(2):713-22. doi: 10.1084/jem.178.2.713. J Exp Med. 1993. PMID: 8393480 Free PMC article. - Thermodynamics of T-cell receptor-peptide/MHC interactions: progress and opportunities.
Armstrong KM, Insaidoo FK, Baker BM. Armstrong KM, et al. J Mol Recognit. 2008 Jul-Aug;21(4):275-87. doi: 10.1002/jmr.896. J Mol Recognit. 2008. PMID: 18496839 Free PMC article. Review. - Structural and dynamic control of T-cell receptor specificity, cross-reactivity, and binding mechanism.
Baker BM, Scott DR, Blevins SJ, Hawse WF. Baker BM, et al. Immunol Rev. 2012 Nov;250(1):10-31. doi: 10.1111/j.1600-065X.2012.01165.x. Immunol Rev. 2012. PMID: 23046120 Review.
Cited by
- Pathogenic epitopes, heterologous immunity and vaccine design.
Welsh RM, Fujinami RS. Welsh RM, et al. Nat Rev Microbiol. 2007 Jul;5(7):555-63. doi: 10.1038/nrmicro1709. Nat Rev Microbiol. 2007. PMID: 17558423 Free PMC article. Review. - Integrating Experiment and Theory to Understand TCR-pMHC Dynamics.
Buckle AM, Borg NA. Buckle AM, et al. Front Immunol. 2018 Dec 7;9:2898. doi: 10.3389/fimmu.2018.02898. eCollection 2018. Front Immunol. 2018. PMID: 30581442 Free PMC article. Review. - CD4 T-cell-mediated heterologous immunity between mycobacteria and poxviruses.
Mathurin KS, Martens GW, Kornfeld H, Welsh RM. Mathurin KS, et al. J Virol. 2009 Apr;83(8):3528-39. doi: 10.1128/JVI.02393-08. Epub 2009 Feb 4. J Virol. 2009. PMID: 19193795 Free PMC article. - Functional recombinant MHC class II molecules and high-throughput peptide-binding assays.
Justesen S, Harndahl M, Lamberth K, Nielsen LL, Buus S. Justesen S, et al. Immunome Res. 2009 May 5;5:2. doi: 10.1186/1745-7580-5-2. Immunome Res. 2009. PMID: 19416502 Free PMC article. - Structural reorganization and the cooperative binding of single-stranded telomere DNA in Sterkiella nova.
Buczek P, Horvath MP. Buczek P, et al. J Biol Chem. 2006 Dec 29;281(52):40124-34. doi: 10.1074/jbc.M607749200. Epub 2006 Nov 2. J Biol Chem. 2006. PMID: 17082188 Free PMC article.
References
- Saito T, Watanabe N. Crit Rev Immunol. 1998;18:359–370. - PubMed
- Bhardwaj V, Kumar V, Geysen H M, Sercarz E E. J Immunol. 1993;151:5000–5010. - PubMed
- Evavold B D, Sloan-Lanchaster J, Wilson K J, Rothbard J B, Allen P M. Immunity. 1995;2:655–663. - PubMed
- Davis M M, Boniface J J, Reich Z, Lyons D, Hampl J, Arden B, Chien Y. Annu Rev Immunol. 1998;16:523–544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials