High-throughput isolation of Caenorhabditis elegans deletion mutants - PubMed (original) (raw)
J M Spoerke, E L Mulligan, J Chen, B Reardon, B Westlund, L Sun, K Abel, B Armstrong, G Hardiman, J King, L McCague, M Basson, R Clover, C D Johnson
Affiliations
- PMID: 10508845
- PMCID: PMC310813
- DOI: 10.1101/gr.9.9.859
High-throughput isolation of Caenorhabditis elegans deletion mutants
L X Liu et al. Genome Res. 1999 Sep.
Abstract
The nematode Caenorhabditis elegans is the first animal whose genome is completely sequenced, providing a rich source of gene information relevant to metazoan biology and human disease. This abundant sequence information permits a broad-based gene inactivation approach in C. elegans, in which chemically mutagenized nematode populations are screened by PCR for deletion mutations in a specific targeted gene. By handling mutagenized worm growth, genomic DNA templates, PCR screens, and mutant recovery all in 96-well microtiter plates, we have scaled up this approach to isolate deletion mutations in >100 genes to date. Four chemical mutagens, including ethyl methane sulfonate, ethlynitrosourea, diepoxyoctane, and ultraviolet-activated trimethylpsoralen, induced detectable deletions at comparable frequencies. The deletions averaged approximately 1400 bp in size when using a approximately 3 kb screening window. The vast majority of detected deletions removed portions of one or more exons, likely resulting in loss of gene function. This approach requires only the knowledge of a target gene sequence and a suitable mutagen, and thus provides a scalable systematic approach to gene inactivation for any organism that can be handled in high density arrays.
Figures
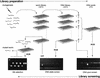
Figure 1
Schematic of mutant library construction and screening. F1 larvae of chemically mutagenized P0 animals are cultured in microtiter arrays to produce F2 larvae, constituting the worm library. An aliquot of worms from each well is removed and processed into a corresponding genomic DNA array; simultaneously, the 96 samples from each plate are pooled and processed to produce genomic DNA plate pools. The DNA plate pools are screened by nested PCR in 96-well plates for deletion amplicons, which identify a positive worm library plate Y. After confirmation by quadruplicate PCR (not illustrated), the genomic DNA array for that specific library plate is similarly screened to identify the specific well. The corresponding well is then retrieved from the frozen mutant worm library and thawed. Individual live worms are allowed to have self-progeny in microtiter wells, and aliquots of each well are tested by PCR to determine which of the thawed worms contain the deletion (see Methods for details).
Figure 2
Deletion size by mutagen, depicting those deletions selected with a PCR screening window of 2800–3400 bp. Not shown are a small number of larger (>3000 bp) deletions identified with PCR primers placed 4–5 kb apart in the target region, as well as smaller (<500 bp) deletions fortuitously detected on screening of specific library plates. Horizontal bar, average deletion size for each mutagen.
Figure 3
Schematics of representative deletions. (a) The deletion of daf-18 PTEN removes a portion of exon 3, which encodes the distal portion of the tensin homology domain and the catalytic phosphatase domain known to be required for lipid phosphatase activity (Gil et al. 1999). (b) The sel-12 deletion removes exons 3–6, which encode predicted transmembrane domains necessary for proper protein topology (Westlund 1998). (c) The cdc-25 deletion removes the entire ORF.
Similar articles
- Isolation of Caenorhabditis elegans gene knockouts by PCR screening of chemically mutagenized libraries.
Lesa GM. Lesa GM. Nat Protoc. 2006;1(5):2231-40. doi: 10.1038/nprot.2006.345. Nat Protoc. 2006. PMID: 17406462 - Reverse genetics by chemical mutagenesis in Caenorhabditis elegans.
Jansen G, Hazendonk E, Thijssen KL, Plasterk RH. Jansen G, et al. Nat Genet. 1997 Sep;17(1):119-21. doi: 10.1038/ng0997-119. Nat Genet. 1997. PMID: 9288111 - Isolation of C. elegans deletion mutants following ENU mutagenesis and thermostable restriction enzyme PCR screening.
Huang CG, Agre P, Strange K, Lamitina T. Huang CG, et al. Mol Biotechnol. 2006 Jan;32(1):83-6. doi: 10.1385/MB:32:1:083. Mol Biotechnol. 2006. PMID: 16382185 - C. elegans deletion mutant screening.
Barstead RJ, Moerman DG. Barstead RJ, et al. Methods Mol Biol. 2006;351:51-8. doi: 10.1385/1-59745-151-7:51. Methods Mol Biol. 2006. PMID: 16988425 Review. - Towards a mutation in every gene in Caenorhabditis elegans.
Moerman DG, Barstead RJ. Moerman DG, et al. Brief Funct Genomic Proteomic. 2008 May;7(3):195-204. doi: 10.1093/bfgp/eln016. Epub 2008 Apr 16. Brief Funct Genomic Proteomic. 2008. PMID: 18417533 Review.
Cited by
- Redundant localization mechanisms of RIM and ELKS in Caenorhabditis elegans.
Deken SL, Vincent R, Hadwiger G, Liu Q, Wang ZW, Nonet ML. Deken SL, et al. J Neurosci. 2005 Jun 22;25(25):5975-83. doi: 10.1523/JNEUROSCI.0804-05.2005. J Neurosci. 2005. PMID: 15976086 Free PMC article. - Efficient isolation of targeted Caenorhabditis elegans deletion strains using highly thermostable restriction endonucleases and PCR.
Wei A, Yuan A, Fawcett G, Butler A, Davis T, Xu SY, Salkoff L. Wei A, et al. Nucleic Acids Res. 2002 Oct 15;30(20):e110. doi: 10.1093/nar/gnf109. Nucleic Acids Res. 2002. PMID: 12384612 Free PMC article. - Transposase-assisted tagmentation: an economical and scalable strategy for single-worm whole-genome sequencing.
Wang Z, Ke J, Guo Z, Wang Y, Lei K, Wang S, Chen G, Shen Z, Li W, Ou G. Wang Z, et al. G3 (Bethesda). 2024 Jul 8;14(7):jkae094. doi: 10.1093/g3journal/jkae094. G3 (Bethesda). 2024. PMID: 38856093 Free PMC article. - Oligoarray comparative genomic hybridization-mediated mapping of suppressor mutations generated in a deletion-biased mutagenesis screen.
Jones MR, Rose AM, Baillie DL. Jones MR, et al. G3 (Bethesda). 2012 Jun;2(6):657-63. doi: 10.1534/g3.112.002238. Epub 2012 Jun 1. G3 (Bethesda). 2012. PMID: 22690375 Free PMC article. - Efficient target-selected mutagenesis in Caenorhabditis elegans: toward a knockout for every gene.
Cuppen E, Gort E, Hazendonk E, Mudde J, van de Belt J, Nijman IJ, Guryev V, Plasterk RH. Cuppen E, et al. Genome Res. 2007 May;17(5):649-58. doi: 10.1101/gr.6080607. Epub 2007 Apr 6. Genome Res. 2007. PMID: 17416746 Free PMC article.
References
- Ahringer J. Turn to the worm. Curr Opin Genet Dev. 1997;7:410–415. - PubMed
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Anderson P. Mutagenesis. in Caenorhabditis elegans: In: Epstein HF, Shakes DC, editors. Modern biological analysis of an organism. San Diego, CA: Academic Press; 1995. pp. 31–58.
- Birren B, Green ED, Klapholz S, Myers RM, Roskams J.Genome analysis: A laboratory manual. 1997. Volume 1: Analyzing DNA1Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 20–22.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources