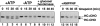The pathway of US11-dependent degradation of MHC class I heavy chains involves a ubiquitin-conjugated intermediate - PubMed (original) (raw)
The pathway of US11-dependent degradation of MHC class I heavy chains involves a ubiquitin-conjugated intermediate
C E Shamu et al. J Cell Biol. 1999.
Abstract
The human cytomegalovirus protein, US11, initiates the destruction of MHC class I heavy chains by targeting them for dislocation from the ER to the cytosol and subsequent degradation by the proteasome. We report the development of a permeabilized cell system that recapitulates US11-dependent degradation of class I heavy chains. We have used this system, in combination with experiments in intact cells, to identify and order intermediates in the US11-dependent degradation pathway. We find that heavy chains are ubiquitinated before they are degraded. Ubiquitination of the cytosolic tail of heavy chain is not required for its dislocation and degradation, suggesting that ubiquitination occurs after at least part of the heavy chain has been dislocated from the ER. Thus, ubiquitination of the heavy chain does not appear to be the signal to start dislocation. Ubiquitinated heavy chains are associated with membrane fractions, suggesting that ubiquitination occurs while the heavy chain is still bound to the ER membrane. Our results support a model in which US11 co-opts the quality control process by which the cell destroys misfolded ER proteins in order to specifically degrade MHC class I heavy chains.
Figures
Figure 1
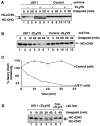
MHC class I heavy chain dislocation and degradation in intact cells and permeabilized cells. (A) The fate of class I heavy chain in intact control and US11 cells. Astrocytoma cells were pulse-labeled for 10 min with [35S]methionine and chased, intact, at 37°C. Samples were taken at the indicated times and cells were lysed with NP-40 lysis buffer. Immunoprecipitations were carried out using rabbit anti–heavy chain serum (αHC). Where indicated, the cells were incubated with the proteasome inhibitor ZL3VS. The bands corresponding to glycosylated heavy chain (HC+CHO) and deglycosylated heavy chain (HC-CHO) are labeled. (B) Heavy chain is degraded in permeabilized US11 cells. US11 cells and control cells were pulse-labeled for 3 min with [35S]methionine, permeabilized, and chased at 37°C for the indicated times. Samples were lysed with NP-40 lysis buffer and class I heavy chain was recovered by immunoprecipitation with αHC serum. (C) The data in B, quantitated on a PhosphorImager. (D) Deglycosylated heavy chain accumulates in US11 cells permeabilized in the presence of the proteasome inhibitor ZL3VS. Cells were labeled, permeabilized, and chased exactly as in B, but in the presence of ZL3VS. Multiple closely spaced heavy chain bands that could often be separated by SDS-PAGE are likely the products of different alleles of MHC class I heavy chain present in the astrocytoma cells.
Figure 2
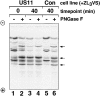
IEF demonstrates that deglycosylated heavy chains accumulate in permeabilized US11 cells. Samples from the experiment shown in Fig. 1 D were treated or mock-treated with PNGase F as indicated, and analyzed by one-dimensional IEF as described (Ploegh 1995). Arrows point to bands corresponding to deglycosylated MHC class I heavy chains. The different HLA gene products migrate with disparate, though characteristic, isoelectric points.
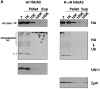
Figure 3
Deglycosylated heavy chain from permeabilized US11 cells is soluble. US11 and control cells were labeled, permeabilized, and chased in the presence of the proteasome inhibitor ZL3VS as described for Fig. 1 D. (A) Fractionation after homogenization. After 30 min of chase at 37°C, the cells were homogenized mechanically and the homogenates were fractionated by centrifugation. Fractions were diluted into NP-40 lysis buffer and immunoprecipitation was carried out with antibodies to HC, transferrin receptor (TfR), β2m, or US11. (B) Fractionation by squeeze-out centrifugation (see Materials and Methods). Total starting material (T), pellet (P), and supernatant (S) fractions are labeled. Denaturing SDS lysates were made and immunoprecipitations were carried out with αHC serum.
Figure 4
Deglycosylated heavy chain is preferentially accessible to protease in permeabilized cells. US11 and control cells were labeled, permeabilized, and chased in the presence of the proteasome inhibitor ZL3VS as described for Fig. 1 D. After 30 min of chase at 37°C, samples from the permeabilization reactions were treated on ice with trypsin, at the final concentrations indicated. Denaturing SDS lysates were made and MHC class I heavy chains were immunoprecipitated with αHC serum. (A) Immunoprecipitation products were analyzed directly by SDS PAGE. Proteolysis produced a glycosylated heavy chain species that lacks its cytoplasmic tail (HCΔtail+CHO). (B) The indicated immunoprecipitation products from (A) were treated (+) or mock-treated (−) with Endo H (New England Biolabs) before SDS PAGE.
Figure 5
Appearance of deglycosylated heavy chain in permeabilized US11 cells requires ATP. (Lanes 1–4) US11 cells were labeled, permeabilized, and chased in the presence of the proteasome inhibitor ZL3VS as described for Fig. 1 D. (Lanes 5–16) Prepared as in lanes 1–4, except that the ATP regenerating system was omitted from the permeabilization buffers and with the additions as indicated. Glucose was at 10 mM, hexokinase was at 0.1 unit/μl, and AMPPNP and magnesium acetate were each at 3 mM. Note that some degradation of heavy chain occurs in the absence of ATP. This degradation is not inhibitable by the addition of proteasome inhibitor (ZL3VS) or standard protease inhibitors (aprotinin, leupeptin, pepstatin, chymostatin, or elastatinal) and has not been characterized further.
Figure 6
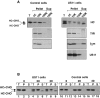
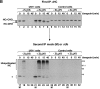
Identification of a ubiquitinated heavy chain intermediate. (A) 125I-ubiquitin labels heavy chains in permeabilized US11 cells (lanes 2–5). US11 and control cells were incubated in solution in the presence or absence of ZL3VS for 1 h. Cells were then incubated with permeabilization buffer containing 125I-labeled ubiquitin. After incubation at 37°C for 10 min, NP-40 lysates were made and subjected to immunoprecipitation with αHC serum followed by SDS PAGE analysis. The mock permeabilization reaction (lane 4) contained no digitonin. [35S]methionine-labeled heavy chain immunoprecipitates from another experiment were run alongside the 125I-labeled samples (lane 1). The migration of molecular mass markers is indicated at the left of the gel. This figure is a composite of nonconsecutive lanes from a single exposure of one gel. (B) Ubiquitinated heavy chain intermediates in 35S-labeled US11 cells in vivo. US11 and control cells were pulse-labeled with [35S]methionine and chased intact at 37°C in the absence or presence of ZL3VS as described for Fig. 1 A. Denaturing SDS lysates were made from samples taken at each timepoint and these were diluted with NP-40 buffer. A first immunoprecipitation was carried out with αHC serum, followed by incubation with Staph A. Bound material was eluted with SDS and one-third of each sample was analyzed directly by SDS PAGE and autoradiography (lanes 1–16). The remaining two thirds of each sample was diluted into NP-40 buffer and reimmunoprecipitated with anti-ubiquitin serum (αUb) or mock-immunoprecipitated (no serum added, M) before SDS PAGE (lanes 17–32). Background bands that precipitate with Staph A alone are identified by the mock immunoprecipitations (*). The migration of molecular mass markers is indicated at the right of the lower gel. Exposure time of the gel for lanes 1–16 is 20 h, and for lanes 17–32 is 30 d.
Figure 6
Identification of a ubiquitinated heavy chain intermediate. (A) 125I-ubiquitin labels heavy chains in permeabilized US11 cells (lanes 2–5). US11 and control cells were incubated in solution in the presence or absence of ZL3VS for 1 h. Cells were then incubated with permeabilization buffer containing 125I-labeled ubiquitin. After incubation at 37°C for 10 min, NP-40 lysates were made and subjected to immunoprecipitation with αHC serum followed by SDS PAGE analysis. The mock permeabilization reaction (lane 4) contained no digitonin. [35S]methionine-labeled heavy chain immunoprecipitates from another experiment were run alongside the 125I-labeled samples (lane 1). The migration of molecular mass markers is indicated at the left of the gel. This figure is a composite of nonconsecutive lanes from a single exposure of one gel. (B) Ubiquitinated heavy chain intermediates in 35S-labeled US11 cells in vivo. US11 and control cells were pulse-labeled with [35S]methionine and chased intact at 37°C in the absence or presence of ZL3VS as described for Fig. 1 A. Denaturing SDS lysates were made from samples taken at each timepoint and these were diluted with NP-40 buffer. A first immunoprecipitation was carried out with αHC serum, followed by incubation with Staph A. Bound material was eluted with SDS and one-third of each sample was analyzed directly by SDS PAGE and autoradiography (lanes 1–16). The remaining two thirds of each sample was diluted into NP-40 buffer and reimmunoprecipitated with anti-ubiquitin serum (αUb) or mock-immunoprecipitated (no serum added, M) before SDS PAGE (lanes 17–32). Background bands that precipitate with Staph A alone are identified by the mock immunoprecipitations (*). The migration of molecular mass markers is indicated at the right of the lower gel. Exposure time of the gel for lanes 1–16 is 20 h, and for lanes 17–32 is 30 d.
Figure 7
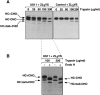
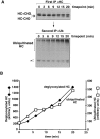
Ubiquitin-conjugated and deglycosylated heavy chains accumulate at the same rate. (A) US11 cells were labeled, permeabilized, and chased in the presence of the proteasome inhibitor ZL3VS as described for Fig. 1 D. αHC immunoprecipitates of each lysate were split and part was reimmunoprecipitated with αUb antibodies, as described for the experiments shown in Fig. 6 B. Nonspecific bands that precipitate with Staph A alone are identified by the asterisk. Note that the exposure of the αHC gel is 12 h whereas the exposure of the αUb gel is 3 wk. (B) The data in A were quantitated on a PhosphorImager and graphed. Note the different scales on the two y axes.
Figure 8
The majority of ubiquitinated heavy chain is not glycosylated. (A) US11 cells were labeled with 3H-mannose in the presence (lane 4) or absence (lane 3) of ZL3VS, denaturing SDS lysates were made, and heavy chains were immunoprecipitated with αHC serum. [35S]methionine-labeled heavy chain immunoprecipitates from another experiment (US11 cells treated with ZL3VS) were run alongside the 3H-mannose–labeled samples (lanes 1 and 2). The asterisk indicates high molecular mass species, most likely ubiquitinated heavy chains, that are immunoprecipitated with αHC serum. The exposure time of this gel is 24 d. (B) US11 cells treated with ZL3VS were labeled with [35S]methionine and chased intact for 0 or 15 min. Denaturing SDS lysates were made and subjected to sequential immunoprecipitation with αHC and αUb serum, as described for Fig. 6 B. One-half of each αUb precipitate was analyzed directly by SDS PAGE (lanes 1 and 2) and the other half was precipitated with Con A–Sepharose, either in the presence (+) or absence (−) of 0.5 M methyl α-
d
-mannopyranoside. Note that the two panels are from a single exposure (4 wk) of the same gel; the lanes were separated to help clarify the experimental procedure. Nonspecific bands that precipitate with Staph A alone are identified by the asterisk. (C) US11 cells from the experiment shown in B but chased intact for 7 min. Con A precipitations were done as in B after immunoprecipitation with αHC serum. The two panels are from a single exposure (3 d) of the same gel.
Figure 9
Fractionation of ubiquitinated heavy chains. (A) US11 cells treated with ZL3VS were permeabilized and chased in the presence of 125I-ubiquitin as in Fig. 6 A. The cells were then homogenized and fractionated by centrifugation as in Fig. 3 A. αHC immunoprecipitates are shown. US11 cells labeled with [35S]methionine but permeabilized without 125I-ubiquitin were fractionated in a parallel experiment; 35S-labeled HC, US11, TfR, and β2m fractionated exactly as in Fig. 3 A (data not shown). The ubiquitinated heavy chain in each fraction was quantitated by PhosphorImager and the results, expressed as a percent of the total amount of ubiquitinated heavy chain, are shown at the bottom of each lane. (B) Squeeze-out fractionation was carried out on 35S-labeled, permeabilized US11 cells treated with ZL3VS as described for Fig. 3 B. MHC class I heavy chains were immunoprecipitated with αHC serum followed by αUb serum, as in Fig. 6 B. The ubiquitinated heavy chain in the pellet and supernatant fractions from the 20-min time point was quantitated by PhosphorImager and the results are shown at the bottom of each lane. Nonspecific bands that precipitate with Staph A alone are identified by the asterisk. Note that the exposure of the αHC immunoprecipitate gel is 56 h, while that of the αUb immunoprecipitate gel is 3 wk. (C and D) Squeeze-out fractionation was carried out on US11 cells in the absence (C) or presence (D) of ZL3VS. In this case, cells were labeled with [35S]methionine and chased intact, and then permeabilized for 10 min on ice in the presence of 0.025% digitonin and the ATP regenerating system before squeeze-out centrifugation. Proteins in each fraction were solubilized with buffer containing 0.5% NP-40 and lysates were prepared for immunoprecipitation as described in Materials and Methods. Sequential immunoprecipitations with αHC serum followed by αUb serum were as in Fig. 6 B. Nonspecific bands that precipitate with Staph A alone are identified by the asterisk. Note that the exposure of the αHC gels in C and D is 16.5 h, while the exposures of the αUb gels are 6 wk (C) and 3 wk (D).
Figure 9
Fractionation of ubiquitinated heavy chains. (A) US11 cells treated with ZL3VS were permeabilized and chased in the presence of 125I-ubiquitin as in Fig. 6 A. The cells were then homogenized and fractionated by centrifugation as in Fig. 3 A. αHC immunoprecipitates are shown. US11 cells labeled with [35S]methionine but permeabilized without 125I-ubiquitin were fractionated in a parallel experiment; 35S-labeled HC, US11, TfR, and β2m fractionated exactly as in Fig. 3 A (data not shown). The ubiquitinated heavy chain in each fraction was quantitated by PhosphorImager and the results, expressed as a percent of the total amount of ubiquitinated heavy chain, are shown at the bottom of each lane. (B) Squeeze-out fractionation was carried out on 35S-labeled, permeabilized US11 cells treated with ZL3VS as described for Fig. 3 B. MHC class I heavy chains were immunoprecipitated with αHC serum followed by αUb serum, as in Fig. 6 B. The ubiquitinated heavy chain in the pellet and supernatant fractions from the 20-min time point was quantitated by PhosphorImager and the results are shown at the bottom of each lane. Nonspecific bands that precipitate with Staph A alone are identified by the asterisk. Note that the exposure of the αHC immunoprecipitate gel is 56 h, while that of the αUb immunoprecipitate gel is 3 wk. (C and D) Squeeze-out fractionation was carried out on US11 cells in the absence (C) or presence (D) of ZL3VS. In this case, cells were labeled with [35S]methionine and chased intact, and then permeabilized for 10 min on ice in the presence of 0.025% digitonin and the ATP regenerating system before squeeze-out centrifugation. Proteins in each fraction were solubilized with buffer containing 0.5% NP-40 and lysates were prepared for immunoprecipitation as described in Materials and Methods. Sequential immunoprecipitations with αHC serum followed by αUb serum were as in Fig. 6 B. Nonspecific bands that precipitate with Staph A alone are identified by the asterisk. Note that the exposure of the αHC gels in C and D is 16.5 h, while the exposures of the αUb gels are 6 wk (C) and 3 wk (D).
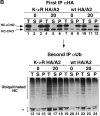
Figure 10
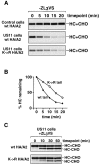
Ubiquitination of the heavy chain cytoplasmic tail is not required for its dislocation from the ER. The stability of HA-tagged MHC class I allele A2 (HA/A2), with either a wt cytoplasmic tail (wt) or with a mutant cytoplasmic tail (K→R), was analyzed in US11 and control cells. Astrocytoma cells stably expressing the HA/A2 constructs were pulse-labeled and chased in the absence (A) or in the presence (C) of ZL3VS, and NP-40 lysates were made as described for Fig. 1 A. HA-tagged heavy chains were immunoprecipitated specifically using monoclonal antibody 12CA5. (B) The data in A were quantitated by PhosphorImager.
Figure 11
HA-tagged heavy chains with either wt or K→R mutant cytoplasmic tails are ubiquitinated. Squeeze-out fractionation, as described for Fig. 9C and Fig. D, was carried out on US11 cells expressing K→R HA/A2 or wt HA/A2. Sequential immunoprecipitation was done on the fractions as in Fig. 6 B, except that the monoclonal antibody 12CA5 was used in the first immunoprecipitation to isolate HA-tagged heavy chains. Experiments were carried out in the absence (A) or presence (B) of ZL3VS. Note that the exposure time of the αHA gels in these panels is 7.5 h and of the αUb gels is 1 wk.
Figure 11
HA-tagged heavy chains with either wt or K→R mutant cytoplasmic tails are ubiquitinated. Squeeze-out fractionation, as described for Fig. 9C and Fig. D, was carried out on US11 cells expressing K→R HA/A2 or wt HA/A2. Sequential immunoprecipitation was done on the fractions as in Fig. 6 B, except that the monoclonal antibody 12CA5 was used in the first immunoprecipitation to isolate HA-tagged heavy chains. Experiments were carried out in the absence (A) or presence (B) of ZL3VS. Note that the exposure time of the αHA gels in these panels is 7.5 h and of the αUb gels is 1 wk.
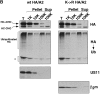
Figure 12
Membrane association of ubiquitinated K→R HA/A2. US11 cells expressing wt or K→R HA/A2 were labeled for 10 min and then homogenized mechanically, without an intervening chase period. Homogenates were fractionated by centrifugation as in Fig. 3 A. Proteins in each fraction were solubilized with buffer containing 0.5% NP-40 and lysates were prepared for immunoprecipitation as described in Materials and Methods. Ubiquitinated, HA-tagged heavy chains were isolated by immunoprecipitating first with the 12CA5 monoclonal antibody, followed by reimmunoprecipitation with αUb serum. Different fractions were also immunoprecipitated for US11 and β2m. Experiments were carried out in the absence (A) or presence (B) of ZL3VS. Nonspecific bands that precipitate with Staph A alone are identified by the asterisk. Note that the exposure of the αHA gel in each panel is 27 h whereas the exposure of the αUb gels is 3 wk (A) or 10 d (B).
Figure 12
Membrane association of ubiquitinated K→R HA/A2. US11 cells expressing wt or K→R HA/A2 were labeled for 10 min and then homogenized mechanically, without an intervening chase period. Homogenates were fractionated by centrifugation as in Fig. 3 A. Proteins in each fraction were solubilized with buffer containing 0.5% NP-40 and lysates were prepared for immunoprecipitation as described in Materials and Methods. Ubiquitinated, HA-tagged heavy chains were isolated by immunoprecipitating first with the 12CA5 monoclonal antibody, followed by reimmunoprecipitation with αUb serum. Different fractions were also immunoprecipitated for US11 and β2m. Experiments were carried out in the absence (A) or presence (B) of ZL3VS. Nonspecific bands that precipitate with Staph A alone are identified by the asterisk. Note that the exposure of the αHA gel in each panel is 27 h whereas the exposure of the αUb gels is 3 wk (A) or 10 d (B).
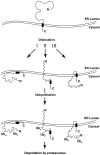
Figure 13
A tentative model for US11-dependent MHC class I heavy chain dislocation and degradation. Arrows I, II, and III indicate three possibilities for how the heavy chain might be associated with the membrane while it is ubiquitinated. Ubn indicates poly-ubiquitin chains and the black box indicates the transmembrane domain of MHC class I heavy chain molecule. See text for details.
Similar articles
- Ubiquitination is essential for human cytomegalovirus US11-mediated dislocation of MHC class I molecules from the endoplasmic reticulum to the cytosol.
Kikkert M, Hassink G, Barel M, Hirsch C, van der Wal FJ, Wiertz E. Kikkert M, et al. Biochem J. 2001 Sep 1;358(Pt 2):369-77. doi: 10.1042/0264-6021:3580369. Biochem J. 2001. PMID: 11513735 Free PMC article. - TRAM1 participates in human cytomegalovirus US2- and US11-mediated dislocation of an endoplasmic reticulum membrane glycoprotein.
Oresic K, Ng CL, Tortorella D. Oresic K, et al. J Biol Chem. 2009 Feb 27;284(9):5905-14. doi: 10.1074/jbc.M807568200. Epub 2009 Jan 2. J Biol Chem. 2009. PMID: 19121997 Free PMC article. - Amino acid composition of alpha1/alpha2 domains and cytoplasmic tail of MHC class I molecules determine their susceptibility to human cytomegalovirus US11-mediated down-regulation.
Barel MT, Pizzato N, van Leeuwen D, Bouteiller PL, Wiertz EJ, Lenfant F. Barel MT, et al. Eur J Immunol. 2003 Jun;33(6):1707-16. doi: 10.1002/eji.200323912. Eur J Immunol. 2003. PMID: 12778489 - The HCMV gene products US2 and US11 target MHC class I molecules for degradation in the cytosol.
van der Wal FJ, Kikkert M, Wiertz E. van der Wal FJ, et al. Curr Top Microbiol Immunol. 2002;269:37-55. doi: 10.1007/978-3-642-59421-2_3. Curr Top Microbiol Immunol. 2002. PMID: 12224515 Review. - Identifying the ERAD ubiquitin E3 ligases for viral and cellular targeting of MHC class I.
van den Boomen DJ, Lehner PJ. van den Boomen DJ, et al. Mol Immunol. 2015 Dec;68(2 Pt A):106-11. doi: 10.1016/j.molimm.2015.07.005. Epub 2015 Jul 22. Mol Immunol. 2015. PMID: 26210183 Free PMC article. Review.
Cited by
- Getting across the cell membrane: an overview for small molecules, peptides, and proteins.
Yang NJ, Hinner MJ. Yang NJ, et al. Methods Mol Biol. 2015;1266:29-53. doi: 10.1007/978-1-4939-2272-7_3. Methods Mol Biol. 2015. PMID: 25560066 Free PMC article. Review. - Battle between Host Immune Cellular Responses and HCMV Immune Evasion.
Manandhar T, Hò GT, Pump WC, Blasczyk R, Bade-Doeding C. Manandhar T, et al. Int J Mol Sci. 2019 Jul 24;20(15):3626. doi: 10.3390/ijms20153626. Int J Mol Sci. 2019. PMID: 31344940 Free PMC article. Review. - Feeling manipulated: cytomegalovirus immune manipulation.
Miller-Kittrell M, Sparer TE. Miller-Kittrell M, et al. Virol J. 2009 Jan 9;6:4. doi: 10.1186/1743-422X-6-4. Virol J. 2009. PMID: 19134204 Free PMC article. - Antigen presentation and the ubiquitin-proteasome system in host-pathogen interactions.
Loureiro J, Ploegh HL. Loureiro J, et al. Adv Immunol. 2006;92:225-305. doi: 10.1016/S0065-2776(06)92006-9. Adv Immunol. 2006. PMID: 17145306 Free PMC article. Review. - Polyubiquitination is required for US11-dependent movement of MHC class I heavy chain from endoplasmic reticulum into cytosol.
Shamu CE, Flierman D, Ploegh HL, Rapoport TA, Chau V. Shamu CE, et al. Mol Biol Cell. 2001 Aug;12(8):2546-55. doi: 10.1091/mbc.12.8.2546. Mol Biol Cell. 2001. PMID: 11514634 Free PMC article.
References
- Baboshina O.V., Haas A.L. Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2EPF and RAD6 are recognized by 26 S proteasome subunit 5. J. Biol. Chem. 1996;271:2823–2831. - PubMed
- Balch W.E., Rothman J.E. Characterization of protein transport between successive compartments of the Golgi apparatusasymmetric properties of donor and acceptor activities in a cell-free system. Arch. Biochem. Biophys. 1985;240:413–425. - PubMed
- Banerjee A., Gregori L., Xu Y., Chau V. The bacterially expressed yeast CDC34 gene product can undergo autoubiquitination to form a multiubiquitin chain-linked protein. J. Biol. Chem. 1993;268:5668–5675. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials