High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology - PubMed (original) (raw)
High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology
M D Krawchuk et al. Yeast. 1999.
Abstract
Bähler et al.(1998) recently described a PCR-based system for the deletion, tagging and overexpression of endogenous genes in the fission yeast Schizosaccharomyces pombe. A small set of PCR primers can be used to generate gene-targeting substrates from each of several modules that differ in the selectable marker (ura4(+) or kanMX6), the presence or absence of specific epitope tags (HA, Myc, GST or GFP), the position in which the epitopes will be inserted (C- or N-terminal), and the presence or absence of a regulatable promoter (the nmt1 promoter). This is a straightforward and powerful system: nine different genes were C-terminal tagged at an average efficiency of 73%, using primers producing only 60-81 bp of homology. In contrast, when studying three transcriptionally-silent genes (rec8(+), rec10(+) and rec11(+)) we obtained an average homologous integration efficiency of 4% for 12 targeting constructs when using primers that contained 80 bp of homology. By using a PCR-based increase in the amount of flanking homology to >/=250 bp, we obtained homologous integration efficiencies of up to 100%. Thus, loci of S. pombe that are refractory to gene targeting when using short tracts of homology can be readily modified by increasing the extent of homology flanking the targeting modules. This straightforward and cost-effective approach might therefore be the one of choice for the modification of S. pombe loci in general and of targeting-refractory loci in particular.
Copyright 1999 John Wiley & Sons, Ltd.
Figures
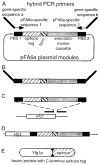
Figure 1
Use of heterologous modules for PCR-based gene targeting in S. pombe. The strategy for placing an epitope tag at the C-terminal codon of a gene is shown, but the same type of approach can be used for N-terminal tagging or gene disruption (Bähler et al., 1998). The schematic diagrams are not to scale. (A) The pFA6a plasmid derivatives (Wach, 1996; Bähler et al., 1998; Longtine et al., 1998) contain two primer binding sites (PBS). PBS 1 encodes a linker polypeptide that is in-frame with one of several modules encoding various different epitope tags. A transcription terminator (term.) is included in most constructs. Also included between the two PBS is a selectable marker cassette containing either the S. pombe ura4+ gene or the kanMX6 gene and all the regulatory sequences required for expression in S. pombe. Hybrid PCR primers are designed to contain approximately 80 nucleotides of homology to the target locus and 20 nucleotides of homology to the PBS. The left-most primer is designed to place the 3′ end of the target locus coding region in-frame with PBS 1 and the epitope module. Because of the modular nature of the pFA6a templates, a single pair of gene-specific primers may be used for each of the modules. (B) PCR amplification generates a gene targeting fragment flanked on each side by about 80 bp of homology to the target locus. (C) Upon transformation, some of the DNA fragments align with the target locus by virtue of this flanking homology. (D) Homologous gene targeting and non-homologous, random integration (not shown) can each give rise to selectable transformants. The type of integration may be readily distinguished by PCR analysis of DNA from transformants. (E) Expression of the modified locus from its endogenous promoter produces a polypeptide that contains the desired epitope tag on its C-terminus.
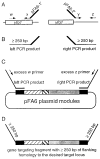
Figure 2
Strategy for a PCR-based increase in the amount of flanking DNA with homology to the target locus. The schematic diagrams are not to scale. (A) Two PCR reactions are configured to amplify independently ≥250 bp of the target locus flanking the desired site of integration. In each case, one of the primers (w or z) is homologous only to the target locus and one of the primers is a hybrid primer containing both gene-specific (x or y) and pFA6a-specific (_pFA6a-1_′ or _pFA6a-2_′) DNA sequence homology. One of the primers (_x, pFA6a-1_′) is designed to place the 3′ end of the target locus coding region in-frame with PBS 1 and the epitope module. (B) The two independent PCR reactions generate long blocks of homology to the target locus (‘left PCR product’ and ‘right PCR product’). (C) The left and right PCR products are subsequently mixed with pFA6a template and an excess of the two outermost, locus-specific PCR primers (w and z). (D) PCR amplification generates a gene targeting fragment flanked on each side by ≥250 bp of homology to the target locus. Transformation and screening of transformants is then as described in Fig. 1C–E.
Similar articles
- Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe.
Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Bähler J, et al. Yeast. 1998 Jul;14(10):943-51. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. Yeast. 1998. PMID: 9717240 - New cassettes for single-step drug resistance and prototrophic marker switching in fission yeast.
Lorenz A. Lorenz A. Yeast. 2015 Dec;32(12):703-10. doi: 10.1002/yea.3097. Epub 2015 Sep 17. Yeast. 2015. PMID: 26305038 - Scarless Gene Tagging with One-Step Transformation and Two-Step Selection in Saccharomyces cerevisiae and Schizosaccharomyces pombe.
Landgraf D, Huh D, Hallacli E, Lindquist S. Landgraf D, et al. PLoS One. 2016 Oct 13;11(10):e0163950. doi: 10.1371/journal.pone.0163950. eCollection 2016. PLoS One. 2016. PMID: 27736907 Free PMC article. - Choosing and using Schizosaccharomyces pombe plasmids.
Siam R, Dolan WP, Forsburg SL. Siam R, et al. Methods. 2004 Jul;33(3):189-98. doi: 10.1016/j.ymeth.2003.11.013. Methods. 2004. PMID: 15157885 Review.
Cited by
- Discovery of genes involved in mitosis, cell division, cell wall integrity and chromosome segregation through construction of Schizosaccharomyces pombe deletion strains.
Chen JS, Beckley JR, Ren L, Feoktistova A, Jensen MA, Rhind N, Gould KL. Chen JS, et al. Yeast. 2016 Sep;33(9):507-17. doi: 10.1002/yea.3172. Epub 2016 Jun 29. Yeast. 2016. PMID: 27168121 Free PMC article. - Characterization of nuclear pore complex components in fission yeast Schizosaccharomyces pombe.
Asakawa H, Yang HJ, Yamamoto TG, Ohtsuki C, Chikashige Y, Sakata-Sogawa K, Tokunaga M, Iwamoto M, Hiraoka Y, Haraguchi T. Asakawa H, et al. Nucleus. 2014 Mar-Apr;5(2):149-62. doi: 10.4161/nucl.28487. Epub 2014 Mar 12. Nucleus. 2014. PMID: 24637836 Free PMC article. - Multistep regulation of protein kinase A in its localization, phosphorylation and binding with a regulatory subunit in fission yeast.
Gupta DR, Paul SK, Oowatari Y, Matsuo Y, Kawamukai M. Gupta DR, et al. Curr Genet. 2011 Oct;57(5):353-65. doi: 10.1007/s00294-011-0354-2. Epub 2011 Aug 31. Curr Genet. 2011. PMID: 21879336 - Featured organism: Schizosaccharomyces pombe, the fission yeast.
Wixon J. Wixon J. Comp Funct Genomics. 2002;3(2):194-204. doi: 10.1002/cfg.92. Comp Funct Genomics. 2002. PMID: 18628834 Free PMC article. - Nsk1 ensures accurate chromosome segregation by promoting association of kinetochores to spindle poles during anaphase B.
Buttrick GJ, Meadows JC, Lancaster TC, Vanoosthuyse V, Shepperd LA, Hoe KL, Kim DU, Park HO, Hardwick KG, Millar JB. Buttrick GJ, et al. Mol Biol Cell. 2011 Dec;22(23):4486-502. doi: 10.1091/mbc.E11-07-0608. Epub 2011 Sep 30. Mol Biol Cell. 2011. PMID: 21965289 Free PMC article.
References
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
- Grallert B, Nurse P, Patterson TE. A study of integrative transformation in Schizosaccharomyces pombe. Mol Gen Genet. 1993;238:26–32. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials