Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus - PubMed (original) (raw)
Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus
A Fassati et al. J Virol. 1999 Nov.
Abstract
To examine the early events in the life cycle of Moloney murine leukemia virus (MoMLV), we analyzed the intracellular complexes mediating reverse transcription. Partial purification of the reverse transcription complexes (RTCs) by equilibrium density fractionation and velocity sedimentation indicated that three distinct species of intracellular complexes are formed shortly after cell infection. Only one of these species is able to start and complete reverse transcription in the cell cytoplasm. This RTC is composed of at least the viral genome, capsid, integrase, and reverse transcriptase proteins. The RTC becomes permeable to micrococcal nuclease but not to antibodies. Shortly after initiation of reverse transcription, the viral strong stop DNA within the RTC is protected from nuclease digestion. The sedimentation velocity of the RTC decreases during reverse transcription. After entry into the nucleus, most capsid proteins are lost from the RTC and its sedimentation velocity decreases further.
Figures
FIG. 1
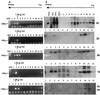
PCR and Western blot analyses of cytoplasmic and nuclear extracts after equilibrium density fractionation. NIH 3T3 fibroblasts were infected with a MoMLV-based vector; cell extracts were prepared 1, 2, 4, and 7 h postinfection, loaded on a 20 to 70% linear sucrose gradient, and centrifuged at 4°C for 20 h at 35,000 rpm in a Beckman SW55 rotor. After centrifugation, gradients were collected in 12 fractions and analyzed. Arrows indicate the direction of the gradient from the lowest (top) to the highest (bottom) density. (A) PCR analyses of the equilibrium density fractions using primers specific for the strong-stop DNA (expected band size is 145 bp). The density of the fraction containing the peak of the viral DNA is indicated for each time point. MW, DNA molecular weight standards. Lanes 1 to 12 correspond to fractions 1 to 12. The rapidly migrating bands are PCR artifacts. (B) The same fractions were precipitated in 10% trichloroacetic acid and analyzed by Western blotting using goat polyclonal antibodies against whole MLV. Purified virus (Virus) and infected NIH 3T3 cells (3T3+) were used as positive controls. Uninfected 3T3 cells (3T3−) were used as negative controls.
FIG. 2
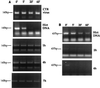
Western blot analyses of equilibrium density fractions containing cytoplasmic extracts collected 4 h postinfection (A) or nuclear extracts collected 7 h postinfection (B). Following SDS-polyacrylamide gel electrophoresis and protein transfer, the polyvinylidene difluoride membrane was probed with the goat polyclonal antibodies against MLV CA or the chicken polyclonal antibodies against MoMLV IN. Lane 1, purified virus; lane 2, uninfected NIH 3T3 cells; lane 3, infected NIH 3T3 cells before equilibrium density fractionation; lane 4, molecular weight standards; lanes 5 to 16, fractions 1 to 12 of the density gradient. The arrow indicates the direction of the gradient from the lowest (top) to the highest (bottom) density.
FIG. 3
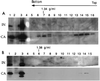
PCR analyses of equilibrium density fractions containing cell extracts collected 1, 2, 4, and 7 h postinfection. (A) Selected fractions from the same equilibrium density gradients shown in Fig. 1 were subjected to PCR using primers specific for the viral elongated minus-strand DNA (expected band size is 437 bp). Fraction numbers are the same as in Fig. 1. Naked viral DNA was used as a positive control (ctr+), and one reaction contained no viral DNA template (ctr−). (B) Fraction 3 of the density gradient containing the nuclear extracts (nuc 3) and fraction 2 of the density gradient containing the cytoplasmic extracts (cyt 2) were subjected to PCR using primers specific for the viral plus-strand DNA (expected band size is 700 bp). Fraction numbers are the same as in Fig. 1. Naked viral DNA was used as a positive control (ctr+), and one reaction contained no viral DNA template (ctr−). The rapidly migrating bands are PCR artifacts.
FIG. 4
Endogenous reverse transcription assay of equilibrium density fractions containing the peak of viral DNA from cytoplasmic extracts collected 1, 2, 4, and 7 h postinfection. Samples diluted 1:10 were incubated with exogenous dNTPs for 6 h at 37°C in the presence (+) or absence (−) of Nonidet P-40 and then subjected to PCR with primers specific for the elongated minus-strand [(−) strand] DNA (expected band size is 437 bp) or the plus-strand [(+) strand] DNA (expected band size is 700 bp). An aliquot of the same fractions was incubated in the absence of dNTPs (no dNTPs). Some samples contained the same amount of cytoplasmic extracts from uninfected cells (ctr−). The rapidly migrating bands are PCR artifacts.
FIG. 5
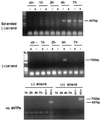
Endogenous reverse transcription assay of equilibrium density gradient fractions containing cytoplasmic extracts collected 1 and 4 h postinfection. Aliquots of the density gradients fractions were incubated at 37°C for 4 h in the presence of dNTPs and Nonidet P-40 and then analyzed by PCR using primers specific for the strong-stop DNA (top; expected band size is 145 bp) or the elongated minus-strand DNA (bottom; expected band size is 437 bp). (A) MW, DNA molecular weight standards; lanes 4 to 11, fractions 4 to 11 of the gradient shown in Fig. 1. (B) Lanes 5 to 12, fractions 5 to 12 of the gradient shown in Fig. 1; mw, DNA molecular weight standards; lane 13, virus subjected to the endogenous reverse transcription assay; lane 14, virus incubated in the absence of dNTPs. Naked DNA was used as positive control for amplification of the elongated minus-strand DNA (ctr+). One reaction contained no viral DNA template (ctr−). The rapidly migrating bands are PCR artifacts.
FIG. 6
Analysis of RTCs by sedimentation velocity. Cytoplasmic extracts collected 1, 2, 4, 7, and 16 h postinfection and nuclear extracts collected 7 h postinfection were subjected to equilibrium density centrifugation. Fractions containing the peak of retroviral DNA were further purified and concentrated in a Centricon filter and centrifuged through a 5 to 20% linear sucrose gradient for 1 h at 23,000 rpm in a Beckman SW55 rotor. Twelve fractions were collected, and the viral DNA in each fraction was detected by PCR using primers specific for the strong-stop DNA (expected band size is 145 bp). The rapidly migrating bands are PCR artifacts.
FIG. 7
The viral strong-stop DNA in the RTC is protected from micrococcal nuclease digestion. Equilibrium density fractions containing the peak of the viral DNA from the cytoplasmic extracts collected 1, 4, and 7 h postinfection were incubated on ice in the presence of micrococcal nuclease and 2 mM CaCl2. The reactions were stopped by addition of 4 mM EGTA at the indicated time points and analyzed by PCR using primers specific for the strong-stop DNA (expected band size is 145 bp) or for the elongated minus-strand DNA (expected band size is 437 bp). Intact virions (CTR virus) incubated in the presence of micrococcal nuclease were used as a negative control. Naked viral DNA was incubated as described above in the presence of micrococcal nuclease and cytoplasmic extracts from uninfected cells (Hirt DNA). The fast-migrating bands are PCR artifacts. To ensure the linearity of amplification, the samples were subjected to two independent amplification rounds of 30 and 40 cycles, respectively.
Similar articles
- Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1.
Fassati A, Goff SP. Fassati A, et al. J Virol. 2001 Apr;75(8):3626-35. doi: 10.1128/JVI.75.8.3626-3635.2001. J Virol. 2001. PMID: 11264352 Free PMC article. - Transcription factor YY1 interacts with retroviral integrases and facilitates integration of moloney murine leukemia virus cDNA into the host chromosomes.
Inayoshi Y, Okino Y, Miyake K, Mizutani A, Yamamoto-Kishikawa J, Kinoshita Y, Morimoto Y, Imamura K, Morshed M, Kono K, Itoh T, Nishijima K, Iijima S. Inayoshi Y, et al. J Virol. 2010 Aug;84(16):8250-61. doi: 10.1128/JVI.02681-09. Epub 2010 Jun 2. J Virol. 2010. PMID: 20519390 Free PMC article. - Abortive reverse transcription by mutants of Moloney murine leukemia virus deficient in the reverse transcriptase-associated RNase H function.
Tanese N, Telesnitsky A, Goff SP. Tanese N, et al. J Virol. 1991 Aug;65(8):4387-97. doi: 10.1128/JVI.65.8.4387-4397.1991. J Virol. 1991. PMID: 1712862 Free PMC article. - Production of a discrete, infectious, double-stranded DNA by reverse transcription in virions of Moloney murine leukemia virus.
Baltimore D, Gilboa E, Rothenberg E, Yoshimura F. Baltimore D, et al. Cold Spring Harb Symp Quant Biol. 1979;43 Pt 2:869-74. doi: 10.1101/sqb.1979.043.01.093. Cold Spring Harb Symp Quant Biol. 1979. PMID: 90578 No abstract available. - Intracytoplasmic maturation of the human immunodeficiency virus type 1 reverse transcription complexes determines their capacity to integrate into chromatin.
Iordanskiy S, Berro R, Altieri M, Kashanchi F, Bukrinsky M. Iordanskiy S, et al. Retrovirology. 2006 Jan 12;3:4. doi: 10.1186/1742-4690-3-4. Retrovirology. 2006. PMID: 16409631 Free PMC article.
Cited by
- The importance of becoming double-stranded: Innate immunity and the kinetic model of HIV-1 central plus strand synthesis.
Poeschla E. Poeschla E. Virology. 2013 Jun 20;441(1):1-11. doi: 10.1016/j.virol.2013.03.010. Epub 2013 Apr 3. Virology. 2013. PMID: 23561461 Free PMC article. Review. - Fates of retroviral core components during unrestricted and TRIM5-restricted infection.
Kutluay SB, Perez-Caballero D, Bieniasz PD. Kutluay SB, et al. PLoS Pathog. 2013 Mar;9(3):e1003214. doi: 10.1371/journal.ppat.1003214. Epub 2013 Mar 7. PLoS Pathog. 2013. PMID: 23505372 Free PMC article. - Restriction of multiple divergent retroviruses by Lv1 and Ref1.
Hatziioannou T, Cowan S, Goff SP, Bieniasz PD, Towers GJ. Hatziioannou T, et al. EMBO J. 2003 Feb 3;22(3):385-94. doi: 10.1093/emboj/cdg042. EMBO J. 2003. PMID: 12554640 Free PMC article.
References
- Alin K, Goff S P. Amino acid substitutions in the CA protein of Moloney murine leukemia virus that block early events in infection. Virology. 1996;222:339–351. - PubMed
- Blain S W, Goff S P. Nuclease activities of Moloney murine leukemia virus reverse transcriptase. J Biol Chem. 1993;268:23585–23592. - PubMed
- Bolognesi D P, Luftig R, Sharper J H. Localization of RNA tumor virus polypeptides. Virology. 1973;56:549–564. - PubMed
- Bowerman B, Brown P O, Bishop J M, Varmus H E. A nucleoprotein complex mediated the integration of retroviral DNA. Genes Dev. 1989;3:469–478. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources