An internal targeting signal directing proteins into the mitochondrial intermembrane space - PubMed (original) (raw)
An internal targeting signal directing proteins into the mitochondrial intermembrane space
K Diekert et al. Proc Natl Acad Sci U S A. 1999.
Abstract
Import of most nucleus-encoded preproteins into mitochondria is mediated by N-terminal presequences and requires a membrane potential and ATP hydrolysis. Little is known about the chemical nature and localization of other mitochondrial targeting signals or of the mechanisms by which they facilitate membrane passage. Mitochondrial heme lyases lack N-terminal targeting information. These proteins are localized in the intermembrane space and are essential for the covalent attachment of heme to c type cytochromes. For import of heme lyases, the translocase of the mitochondrial outer membrane complex is both necessary and sufficient. Here, we report the identification of the targeting signal of mitochondrial heme lyases in the third quarter of these proteins. The targeting sequence is highly conserved among all known heme lyases. Its chemical character is hydrophilic because of a large fraction of both positively and negatively charged amino acid residues. These features clearly distinguish this signal from classical presequences. When inserted into a cytosolic protein, the targeting sequence directs the fusion protein into the intermembrane space, even in the absence of a membrane potential or ATP hydrolysis. The heme lyase targeting sequence represents the first topogenic signal for energy-independent transport into the intermembrane space and harbors two types of information. It assures accurate recognition and translocation by the translocase of the mitochondrial outer membrane complex, and it is responsible for driving the import reaction by undergoing high-affinity interactions with components of the intermembrane space.
Figures
Figure 1
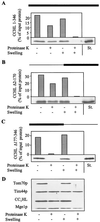
CCHL does not contain essential import information in its N-terminal half. The precursor proteins CCHL (A), CCHL Δ2–170 (B), and CCHL Δ177–346 (C; see Materials and Methods) were synthesized by in vitro transcription and translation in reticulocyte lysate by using [35S]methionine as a label. The black bar on top of each panel represents the region of CCHL present in these proteins; the thin line corresponds to deleted segments. The radiolabeled proteins were added to isolated mitochondria in import buffer (36). After incubation for 10 min at 25°C, mitochondria were reisolated by centrifugation (10 min at 9,000 × g). Mitochondria were resuspended in a small volume of SoH buffer (0.6 M sorbitol_/20 mM Hepes-KOH, pH 7.2). Samples were diluted 10-fold into SoH buffer or water in the presence or absence of proteinase K as indicated. The hypotonic condition results in swelling of the organelles and leads to selective rupture of the outer membrane allowing added proteinase K to degrade proteins exposed in the intermembrane space (D, CC1HL) but not of proteins of the matrix (Tim44p and Mge1p). After incubation for 30 min at 0°C, protease digestion was halted by the addition of PMSF, and proteins were precipitated with trichloroacetic acid. Proteins were separated by SDS/_PAGE, blotted on nitrocellulose, and quantified by PhosphorImager analysis. In addition, an autoradiograph is shown. The material that is not digested after swelling represents aggregated preprotein. The rightmost lanes contain 50% of input preprotein as a standard (St.). Import (i.e., protease-resistant protein relative to bound material) varied by not more than 15% in various experiments.
Figure 2
The third quarter of CCHL contains both necessary and sufficient targeting information. The import of the indicated CCHL mutant proteins was estimated as described in the legend of Fig. 1. The amount of radiolabeled protein was quantitated by PhosphorImager analysis and is given above the autoradiographs relative to the amount of preprotein bound to mitochondria (set to 100%). The rightmost lanes contain 50% of input preprotein (St., standard).
Figure 3
The import information of N. crassa CCHL is contained within two conserved small segments. Import of the indicated CCHL mutant proteins was estimated and analyzed as described in the legend of Fig. 2. St., standard representing 50% of input preprotein.
Figure 4
The targeting sequence of heme lyases can direct a cytosolic protein into the intermembrane space. The indicated regions of N. crassa CCHL (A and B) or of S. cerevisiae CC1HL (C) were inserted into the cytosolic protein DHFR. Import of these fusion proteins or of CC1HL (D) and further analysis of import were performed as described in the legend of Fig. 2. Other parts of the heme lyases did not support the import of the corresponding fusion proteins (not shown). The DHFR sequence is indicated by the hatched boxes; the N. crassa CCHL and S. cerevisiae CC1HL sequences are given as black and grey bars, respectively. St., standard representing 50% of input preprotein.
Figure 5
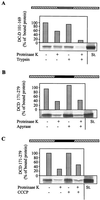
Import mediated by the heme lyase targeting sequence occurs along the authentic heme lyase import pathway. The fusion proteins DCD 171–279 and DC1D 101–169 were imported into isolated mitochondria. (A) The organelles were pretreated with 50 μg_/ml trypsin (27). (B) ATP was hydrolyzed by treatment with 20 units/ml apyrase (ref. ; import reactions contained 20 μM oligomycin and 5 μM carboxy-atractyloside). (C) The membrane potential was depleted by the addition of the uncoupling reagent carbonyl cyanide m-chlorophenylhydrazone (50 μM; CCCP; ref. 26). Under any of these conditions, import of presequence-containing preproteins into the matrix was strongly impaired (not shown). Further treatment of the samples and analysis of import were performed as described in Fig. 2. The result for only one of the heme lyase/_DHFR fusion proteins is shown.
Figure 6
The heme lyase targeting signal encompasses two signature motifs that are highly conserved in these proteins. The sequence alignment of known heme lyases was prepared by using the
multalin
program (54). No bacterial homologues have been identified. The heme lyase targeting signal determined in this study is underlined. The secondary structure of these sequences was predicted by using the
phdsec
program (55) revealing similar results for CCHL and CC1HL proteins. H, α-helical; E, extended β-stranded. Nc, N. crassa; Sp, Schizosaccharomyces pombe; Sc, S. cerevisiae; Ca, Candida albicans; Hs, Homo sapiens; Mm, Mus musculus; Ce, Caenorhabditis elegans.
Similar articles
- Biogenesis of mitochondrial heme lyases in yeast. Import and folding in the intermembrane space.
Steiner H, Zollner A, Haid A, Neupert W, Lill R. Steiner H, et al. J Biol Chem. 1995 Sep 29;270(39):22842-9. doi: 10.1074/jbc.270.39.22842. J Biol Chem. 1995. PMID: 7559417 - Import of cytochrome c heme lyase into mitochondria: a novel pathway into the intermembrane space.
Lill R, Stuart RA, Drygas ME, Nargang FE, Neupert W. Lill R, et al. EMBO J. 1992 Feb;11(2):449-56. doi: 10.1002/j.1460-2075.1992.tb05074.x. EMBO J. 1992. PMID: 1371459 Free PMC article. - Functional independence of the protein translocation machineries in mitochondrial outer and inner membranes: passage of preproteins through the intermembrane space.
Segui-Real B, Kispal G, Lill R, Neupert W. Segui-Real B, et al. EMBO J. 1993 May;12(5):2211-8. doi: 10.1002/j.1460-2075.1993.tb05869.x. EMBO J. 1993. PMID: 8491208 Free PMC article. - Mitochondrial protein import.
Hartl FU, Pfanner N, Nicholson DW, Neupert W. Hartl FU, et al. Biochim Biophys Acta. 1989 Jan 18;988(1):1-45. doi: 10.1016/0304-4157(89)90002-6. Biochim Biophys Acta. 1989. PMID: 2642391 Review. - The mitochondrial import machinery for preproteins.
Rehling P, Wiedemann N, Pfanner N, Truscott KN. Rehling P, et al. Crit Rev Biochem Mol Biol. 2001;36(3):291-336. doi: 10.1080/20014091074200. Crit Rev Biochem Mol Biol. 2001. PMID: 11450972 Review.
Cited by
- Divergent Cytochrome c Maturation System in Kinetoplastid Protists.
Belbelazi A, Neish R, Carr M, Mottram JC, Ginger ML. Belbelazi A, et al. mBio. 2021 May 4;12(3):e00166-21. doi: 10.1128/mBio.00166-21. mBio. 2021. PMID: 33947751 Free PMC article. - An efficient genetic screen in Drosophila to identify nuclear-encoded genes with mitochondrial function.
Liao TS, Call GB, Guptan P, Cespedes A, Marshall J, Yackle K, Owusu-Ansah E, Mandal S, Fang QA, Goodstein GL, Kim W, Banerjee U. Liao TS, et al. Genetics. 2006 Sep;174(1):525-33. doi: 10.1534/genetics.106.061705. Epub 2006 Jul 18. Genetics. 2006. PMID: 16849596 Free PMC article. - Conserved residues of the human mitochondrial holocytochrome c synthase mediate interactions with heme.
Babbitt SE, San Francisco B, Bretsnyder EC, Kranz RG. Babbitt SE, et al. Biochemistry. 2014 Aug 19;53(32):5261-71. doi: 10.1021/bi500704p. Epub 2014 Aug 6. Biochemistry. 2014. PMID: 25054239 Free PMC article. - Ups delivery to the intermembrane space of mitochondria: a novel affinity-driven protein import pathway.
Herrmann JM. Herrmann JM. EMBO J. 2010 Sep 1;29(17):2859-60. doi: 10.1038/emboj.2010.189. EMBO J. 2010. PMID: 20811336 Free PMC article. - Roles for the Rad27 Flap Endonuclease in Mitochondrial Mutagenesis and Double-Strand Break Repair in Saccharomyces cerevisiae.
Nagarajan P, Prevost CT, Stein A, Kasimer R, Kalifa L, Sia EA. Nagarajan P, et al. Genetics. 2017 Jun;206(2):843-857. doi: 10.1534/genetics.116.195149. Epub 2017 Apr 26. Genetics. 2017. PMID: 28450457 Free PMC article.
References
- Neupert W. Annu Rev Biochem. 1997;66:861–915. - PubMed
- Schatz G, Dobberstein B. Science. 1996;271:1519–1526. - PubMed
- Pfanner N, Craig E A, Honlinger A. Annu Rev Cell Dev Biol. 1997;13:25–51. - PubMed
- Roise D, Schatz G. J Biol Chem. 1988;263:4509–4511. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases