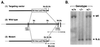Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2 - PubMed (original) (raw)
. 1999 Oct 12;96(21):12114-9.
doi: 10.1073/pnas.96.21.12114.
C P Selby, T Todo, H Niwa, C Thompson, E M Fruechte, K Hitomi, R J Thresher, T Ishikawa, J Miyazaki, J S Takahashi, A Sancar
Affiliations
- PMID: 10518585
- PMCID: PMC18421
- DOI: 10.1073/pnas.96.21.12114
Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2
M H Vitaterna et al. Proc Natl Acad Sci U S A. 1999.
Abstract
Cryptochromes regulate the circadian clock in animals and plants. Humans and mice have two cryptochrome (Cry) genes. A previous study showed that mice lacking the Cry2 gene had reduced sensitivity to acute light induction of the circadian gene mPer1 in the suprachiasmatic nucleus (SCN) and had an intrinsic period 1 hr longer than normal. In this study, Cry1(-/-) and Cry1(-/-)Cry2(-/-) mice were generated and their circadian clocks were analyzed at behavioral and molecular levels. Behaviorally, the Cry1(-/-) mice had a circadian period 1 hr shorter than wild type and the Cry1(-/-)Cry2(-/-) mice were arrhythmic in constant darkness (DD). Biochemically, acute light induction of mPer1 mRNA in the SCN was blunted in Cry1(-/-) and abolished in Cry1(-/-)Cry2(-/-) mice. In contrast, the acute light induction of mPer2 in the SCN was intact in Cry1(-/-) and Cry1(-/-)Cry2(-/-) animals. Importantly, in double mutants, mPer1 expression was constitutively elevated and no rhythmicity was detected in either 12-hr light/12-hr dark or DD, whereas mPer2 expression appeared rhythmic in 12-hr light/12-hr dark, but nonrhythmic in DD with intermediate levels. These results demonstrate that Cry1 and Cry2 are required for the normal expression of circadian behavioral rhythms, as well as circadian rhythms of mPer1 and mPer2 in the SCN. The differential regulation of mPer1 and mPer2 by light in Cry double mutants reveals a surprising complexity in the role of cryptochromes in mammals.
Figures
Figure 1
Targeted disruption of the Cry1 gene. (A) Targeting of the Cry1 locus. The construct (1) was used to target the Cry1 gene (2) in the E14 g2a embryonic stem cell line. Homologous recombination leads to the deletion of a 13-kb genomic region [stippled box in (2)] containing exon sequences encoding the FAD-binding domain. The targeted allele (3) is detected by a probe as shown, with the expected DNA fragment sizes as indicated. Solid boxes, identified coding sequences; SA, Engrail-2 splice acceptor; S, _Sal_I; Xb, _Xba_I; Xh, _Xho_I (restriction sites). (B) Identification of targeted mutants by Southern hybridization. The 2.5-kb mutant and the 6.6-kb wild-type fragments resulting from Xba digestion are indicated.
Figure 2
Genotyping of progeny from Cry1+/−Cry2+/− cross by PCR. The knockouts of both genes each were generated by deleting a segment of the wild-type gene encoding the FAD-binding domain of CRY1 (amino acids 230–549) and of CRY2 (amino acids 349–569) and replacing it with the Neo gene. As shown in the schematic diagram, primers hybridizing to the deleted region were used to detect the wild type and primers hybridizing to the Neo gene were used to detect the mutated genes. The photograph shows results of PCR analysis of wild-type, double heterozygous, and double homozygous mutant mice.
Figure 3
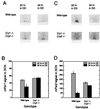
Effects of disruption of the Cry genes on circadian locomotor activity rhythms. (A_–_D) Wheel-running activity records of individual mice, double-plotted according to convention so that each day’s data are represented both to the right and beneath that of the preceding day. Times of activity are represented by black. The animals were kept on a LD12:12 cycle as indicated by the bar above each record and then transferred to constant darkness by allowing lights to go off at the usual time on the day, indicated by an arrow on the right. (A) Activity record of a wild-type mouse. (B) Activity record of a _Cry1_−/− mouse. (C) Activity record of a _Cry2_−/− mouse. (D) Activity record of a _Cry1_−/−_Cry2_−/− mouse. (E) Effects of disruption of the Cry genes on circadian period. The free-running period was estimated by χ2 periodogram from days 1–20 in DD. Means and SEM of each genotype are illustrated. Sample sizes (N) are as follows: wild-type, n = 6; _Cry1_−/−, n = 5; _Cry2_−/−, n = 10; _Cry1_−/−Cry2−/−, n = 4. None of the double homozygotes exhibited significant circadian periodicity and, hence, no period estimates are shown. (F) Loss of circadian rhythmicity was assessed by Fourier analysis. Data from days 1–20 in DD were analyzed by fast Fourier transform (FFT), and power spectral densities for frequencies ranging from 0 to 1 cycles/hr were determined and normalized to a total power (area under the curve) of one. The resultant relative power value peak in the circadian range (18- to 30-hr period or 0.033- 0.055 cycles/hr) was determined for each animal for comparison. The means and SEM of circadian peak values of relative power are plotted for each genotype. Sample sizes are the same as in E.
Figure 4
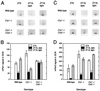
Effects of disruption of the Cry genes on diurnal expression of mPer1 and mPer2. (A) Representative in situ mPer1 signal in the SCN regions of mice of three different genotypes and under three different conditions. (B) Means and ranges in signal values for mPer1 in the SCN by time/light condition and genotype. (C) Representative in situ mPer2 signal in the SCN regions of mice of three different genotypes and under three different conditions. (D) Means and ranges in signal values for mPer2 in the SCN by time/light condition and genotype. n = 2 per genotype per condition.
Figure 5
Effects of disruption of the Cry genes on circadian expression of mPer1 and mPer2. (A) Representative in situ mPer1 signal in the SCN regions of wild-type and _Cry1_−/−_Cry2_−/− mutant mice and at two times in DD. (B) Means and ranges in signal values for mPer1 in the SCN by time and genotype. (C) Representative in situ mPer2 signal in the SCN regions of wild-type and _Cry1_−/−_Cry2_−/− mutant mice and at two times in DD. (D) Means and ranges in signal values for mPer2 in the SCN by time and genotype. n = 3 wild types; two mutants per condition.
Figure 6
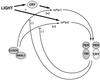
The dual role of cryptochromes in the circadian clock. A model of genetic interactions among cryptochromes and other elements in the circadian autoregulatory loop are shown. A presumed basic feedback loop of positive mPer transcriptional drive of CLOCK-BMAL1 inhibited by PER-TIM is depicted. Cryptochromes appear to mediate light induction of mPer1 but not mPer2 (left side). In addition, the potential for cryptochromes to dimerize with PERs suggests that they may both function in a negative feedback as well (right side).
Similar articles
- Disruption of mCry2 restores circadian rhythmicity in mPer2 mutant mice.
Oster H, Yasui A, van der Horst GT, Albrecht U. Oster H, et al. Genes Dev. 2002 Oct 15;16(20):2633-8. doi: 10.1101/gad.233702. Genes Dev. 2002. PMID: 12381662 Free PMC article. - Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock.
Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, Muijtjens M, Hoeijmakers JH, van der Horst GT. Okamura H, et al. Science. 1999 Dec 24;286(5449):2531-4. doi: 10.1126/science.286.5449.2531. Science. 1999. PMID: 10617474 - Targeted disruption of the mPer3 gene: subtle effects on circadian clock function.
Shearman LP, Jin X, Lee C, Reppert SM, Weaver DR. Shearman LP, et al. Mol Cell Biol. 2000 Sep;20(17):6269-75. doi: 10.1128/MCB.20.17.6269-6275.2000. Mol Cell Biol. 2000. PMID: 10938103 Free PMC article. - Cryptochrome: the second photoactive pigment in the eye and its role in circadian photoreception.
Sancar A. Sancar A. Annu Rev Biochem. 2000;69:31-67. doi: 10.1146/annurev.biochem.69.1.31. Annu Rev Biochem. 2000. PMID: 10966452 Review. - [Molecular mechanisms of biological clock: from molecular rhythms to physiological rhythms].
Okamura H. Okamura H. No To Shinkei. 2003 Jan;55(1):5-11. No To Shinkei. 2003. PMID: 12649895 Review. Japanese. No abstract available.
Cited by
- A Cryptochrome 2 mutation yields advanced sleep phase in humans.
Hirano A, Shi G, Jones CR, Lipzen A, Pennacchio LA, Xu Y, Hallows WC, McMahon T, Yamazaki M, Ptáček LJ, Fu YH. Hirano A, et al. Elife. 2016 Aug 16;5:e16695. doi: 10.7554/eLife.16695. Elife. 2016. PMID: 27529127 Free PMC article. - Mood phenotypes in rodent models with circadian disturbances.
Imamura K, Takumi T. Imamura K, et al. Neurobiol Sleep Circadian Rhythms. 2022 Oct 13;13:100083. doi: 10.1016/j.nbscr.2022.100083. eCollection 2022 Nov. Neurobiol Sleep Circadian Rhythms. 2022. PMID: 36345502 Free PMC article. Review. - Integration of the circadian and stress systems: influence of neuropeptides and implications for alcohol consumption.
Wong CC, Schumann G. Wong CC, et al. J Neural Transm (Vienna). 2012 Oct;119(10):1111-20. doi: 10.1007/s00702-012-0829-4. Epub 2012 May 31. J Neural Transm (Vienna). 2012. PMID: 22648536 Review. - USP2a protein deubiquitinates and stabilizes the circadian protein CRY1 in response to inflammatory signals.
Tong X, Buelow K, Guha A, Rausch R, Yin L. Tong X, et al. J Biol Chem. 2012 Jul 20;287(30):25280-91. doi: 10.1074/jbc.M112.340786. Epub 2012 Jun 5. J Biol Chem. 2012. PMID: 22669941 Free PMC article. - Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β.
Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, Liddle C, Auwerx J, Downes M, Panda S, Evans RM. Cho H, et al. Nature. 2012 Mar 29;485(7396):123-7. doi: 10.1038/nature11048. Nature. 2012. PMID: 22460952 Free PMC article.
References
- Young M. Annu Rev Biochem. 1998;37:135–152. - PubMed
- Green C B. Trends Cell Biol. 1998;8:224–230. - PubMed
- Dunlap J C. Cell. 1999;96:271–290. - PubMed
- Herzog E D, Takahashi J S, Block G D. Nat Neurosci. 1998;1:708–713. - PubMed
- Cashmore A R. J Plant Res. 1998;111:267–270.
Publication types
MeSH terms
Substances
Grants and funding
- GM31082/GM/NIGMS NIH HHS/United States
- R37 GM031082/GM/NIGMS NIH HHS/United States
- R01 GM031082/GM/NIGMS NIH HHS/United States
- AG11412/AG/NIA NIH HHS/United States
- P01 AG011412/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous