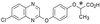XK469, a selective topoisomerase IIbeta poison - PubMed (original) (raw)
XK469, a selective topoisomerase IIbeta poison
H Gao et al. Proc Natl Acad Sci U S A. 1999.
Abstract
XK469 (NSC 697887) is a synthetic quinoxaline phenoxypropionic acid derivative that possesses unusual solid tumor selectivity and activity against multidrug-resistant cancer cells. We report here that XK469 and its S(-) and R(+)-isomers induce reversible protein-DNA crosslinks in mammalian cells. Under protein denaturing conditions, the protein-DNA crosslinks are rendered irreversible and stable to DNA banding by CsCl gradient ultracentrifugation. Several lines of evidence indicate that the primary target of XK469 is topoisomerase IIbeta. Preferential targeting of topoisomerase IIbeta may explain the solid tumor selectivity of XK469 and its analogs because solid tumors, unlike leukemias, often have large populations of cells in the G(1)/G(0) phases of the cell cycle in which topoisomerase IIbeta is high whereas topoisomerase IIalpha, the primary target of many leukemia selective drugs, is low.
Figures
Figure 1
Structure of XK469. The acid form of XK469 is shown, and the asymmetric center is indicated by an asterisk.
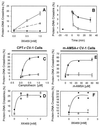
Figure 2
XK469-induced protein-DNA crosslinks to cellular and SV40 DNA. (A) Replicating SV40 genomes were pulse-labeled with3H-dT for 30 min, and the drugs were added 15 min after the start of labeling. Labeling medium with drug was drawn off and the cells were lysed with Hirt lysing fluid. The Hirt supernatant, containing pulse-labeled SV40 DNA, was assayed for protein-SV40 DNA crosslinks. ○, R-isomer; ▵, S-isomer; ▾ R-isomer, proteinase K digested before assay; ⋄, S-isomer, proteinase K digested before assay. Error bars are ±SD derived from four points. (B) Reversal of S(−)XK469-induced protein-DNA crosslinks upon removal of the drug. SV40-infected CV-1 cells were labeled with 3H-dT beginning at t = −30 min. At t = −15 min, the labeling medium in all samples was made 1 mM in S(−)XK469. At t = 0, the labeling medium with XK469 in one set of samples was replaced with labeling medium without drug. At t = 2, 10, and 20 min, samples were harvested by removal of labeling medium and addition of Hirt lysing fluid. SV40 DNA was selectively extracted and measured for protein-DNA crosslinks (see Materials and Methods). (C) Camptothecin-induced topoisomerase I-cellular DNA crosslinks in drug-sensitive parental CV-1 cells (●) and in CPTCV10c22 cells (○) resistant to 1.5 μM camptothecin (CPT-r). (D) S(−)XK469-induced protein-DNA crosslinks in drug-sensitive parental CV-1 cells (●) and in CPTCV10c22 cells (○). (E) _m_-AMSA-induced topoisomerase II-DNA crosslinks in drug-sensitive parental CV-1 cells (●) and in AMCV1 cells (○) resistant to 3 μM_m_-AMSA. (F) S(−)XK469-induced protein-DNA crosslinks in parental CV-1 cells (●) and in AMCV1 (○) cells.
Figure 3
Western blot of topoisomerase II isozymes in AMCV1 cells and band depletion assays for topoisomerase II isozymes in CV-1 cells. (A Right lanes) Western blot analysis of 3 μM_m_-AMSA-resistant AMCV1 cells (AMSAr). Extracts of drug-sensitive parental CV-1 cells and AMCV1 cells were subjected to acrylamide gel electrophoresis in SDS. The separated proteins were transferred to a poly(vinylidene difluoride) membrane and were probed with topoisomerase IIα- or topoisomerase IIβ-specific antibody (Ab). (Left lanes) Band depletion analysis of topoisomerase II isozymes in CV-1 cells treated with VM-26 and XK469. Cells were treated for 15 min with the indicated drugs before extraction. The control was treated with the drug solvent, DMSO. Camptothecin (CPT), a topoisomerase I poison, was included as an additional control. The S-isomer of XK469 was used in this experiment. (B) Band depletion for topoisomerase IIβ isozyme in CV-1 cells treated with_m_-AMSA, XK469, and camptothecin.
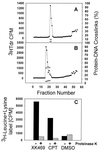
Figure 4
XK469-induced protein-DNA crosslinks stable to DNA banding by cesium chloride ultracentrifugation. GM637d2 human fibroblasts were labeled with 3H-dT (1 μCi/ml, 43 hr). The cells were then either treated with the solvent, DMSO (A), or with 1 mM (±)XK469, acid form (B) for 15 min. The labeling medium was drawn off, and the cells then were lysed by addition of 6 M GuHCl. DNA was sheared by vortexing the GuHCl cell lysate, and the samples then were layered onto CsCl step gradients (see Materials and Methods). After ultracentrifugation and elution of the gradient, 5 μl of each 200 μl fraction was removed for direct scintillation counting. DNA containing fractions from each experiment, identified by tritium incorporation, were dialyzed against TE buffer (10 mM Tris⋅HCl, pH 7.6/1 mM EDTA) to remove CsCl and then were assayed for protein-DNA crosslinks by the GF/C filter assay. Tritiated thymidine incorporation (DNA, ●) is indicated on the left axis of each graph, and protein-DNA crosslinks are indicated on the right axis either with (▿) or without (○) proteinase K predigestion. Fractions were pooled in pairs for the assays, and the value for the pair is indicated for each fraction. (C) Protein labeled with 3H-leucine + lysine also bands with DNA by CsCl ultracentrifugation after exposure to either 1 mM (±)XK469 (acid form) or to 10 μM camptothecin (black bars). Predigestion with proteinase K (light bars) eliminates this protein label associated with the DNA.
Figure 5
Covalent binding of topoisomerase IIα and topoisomerase IIβ to cellular DNA after exposure of cells to topoisomerase poisons. Subconfluent and postconfluent human breast cancer cells (MCF-7) were treated with S(−)XK469, VM-26, m_-AMSA, or the solvent, DMSO, for 15 min and then were lysed with GuHCl as described (see_Materials and Methods). The cellular DNA was banded by CsCl gradient ultracentrifugation, and the CsCl then was removed by dialysis of the pooled DNA fractions. A 30-μg DNA aliquot was removed, and MgCl2 was added to a final concentration of 5 mM. The DNA was digested with protease-free DNase I (Boehringer Mannheim, 0.1 units/ml, 37°C, 1 hr). The aliquot then was applied to a poly(vinylidene difluoride) membrane with a slot blot device. Purified human topoisomerase IIα (TopoGen) also was applied as a control. Blotted proteins were probed with antibodies to human topoisomerase IIα and human topoisomerase IIβ (indicated at the top of each column).
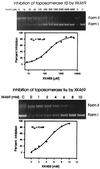
Figure 6
Catalytic inhibition of topoisomerase II isozymes by S(−)XK469. ATP-dependent relaxation of superhelical pBR322 DNA is shown for purified human topoisomerase IIβ (Upper) and purified human topoisomerase IIα (Lower). Inhibition was measured by quantitation of the form I band in the ethidium stained gels. The graph from which the IC50 value was determined is shown below each gel. Controls (C) are pBR322 DNA without added enzyme.
Similar articles
- Cytotoxic mechanism of XK469: resistance of topoisomerase IIbeta knockout cells and inhibition of topoisomerase I.
Snapka RM, Gao H, Grabowski DR, Brill D, Chan KK, Li L, Li GC, Ganapathi R. Snapka RM, et al. Biochem Biophys Res Commun. 2001 Feb 2;280(4):1155-60. doi: 10.1006/bbrc.2001.4249. Biochem Biophys Res Commun. 2001. PMID: 11162648 - DNA sequence specificity for topoisomerase II poisoning by the quinoxaline anticancer drugs XK469 and CQS.
Gao H, Yamasaki EF, Chan KK, Shen LL, Snapka RM. Gao H, et al. Mol Pharmacol. 2003 Jun;63(6):1382-8. doi: 10.1124/mol.63.6.1382. Mol Pharmacol. 2003. PMID: 12761349 - Chloroquinoxaline sulfonamide (NSC 339004) is a topoisomerase IIalpha/beta poison.
Gao H, Yamasaki EF, Chan KK, Shen LL, Snapka RM. Gao H, et al. Cancer Res. 2000 Nov 1;60(21):5937-40. Cancer Res. 2000. PMID: 11085507 - Topoisomerase IIα, rather than IIβ, is a promising target in development of anti-cancer drugs.
Chen W, Qiu J, Shen YM. Chen W, et al. Drug Discov Ther. 2012 Oct;6(5):230-7. Drug Discov Ther. 2012. PMID: 23229142 Review. - DNA and associated targets for drug design.
Hurley LH. Hurley LH. J Med Chem. 1989 Sep;32(9):2027-33. doi: 10.1021/jm00129a001. J Med Chem. 1989. PMID: 2671370 Review. No abstract available.
Cited by
- Nitric oxide inhibits ATPase activity and induces resistance to topoisomerase II-poisons in human MCF-7 breast tumor cells.
Sinha BK, Kumar A, Mason RP. Sinha BK, et al. Biochem Biophys Rep. 2017 Apr 20;10:252-259. doi: 10.1016/j.bbrep.2017.04.011. eCollection 2017 Jul. Biochem Biophys Rep. 2017. PMID: 28955753 Free PMC article. - Natural Compounds as Anticancer Agents Targeting DNA Topoisomerases.
Jain CK, Majumder HK, Roychoudhury S. Jain CK, et al. Curr Genomics. 2017 Feb;18(1):75-92. doi: 10.2174/1389202917666160808125213. Curr Genomics. 2017. PMID: 28503091 Free PMC article. Review. - Lycobetaine acts as a selective topoisomerase II beta poison and inhibits the growth of human tumour cells.
Barthelmes HU, Niederberger E, Roth T, Schulte K, Tang WC, Boege F, Fiebig HH, Eisenbrand G, Marko D. Barthelmes HU, et al. Br J Cancer. 2001 Nov 16;85(10):1585-91. doi: 10.1054/bjoc.2001.2142. Br J Cancer. 2001. PMID: 11720449 Free PMC article. - NK314, a topoisomerase II inhibitor that specifically targets the alpha isoform.
Toyoda E, Kagaya S, Cowell IG, Kurosawa A, Kamoshita K, Nishikawa K, Iiizumi S, Koyama H, Austin CA, Adachi N. Toyoda E, et al. J Biol Chem. 2008 Aug 29;283(35):23711-20. doi: 10.1074/jbc.M803936200. Epub 2008 Jul 2. J Biol Chem. 2008. PMID: 18596031 Free PMC article. - The chemotherapeutic agents XK469 (2-{4-[(7-chloro-2-quinoxalinyl)oxy]phenoxy}propionic acid) and SH80 (2-{4-[(7-bromo-2-quinolinyl)oxy]phenoxy}propionic acid) inhibit cytokinesis and promote polyploidy and induce senescence.
Reiners JJ Jr, Kleinman M, Joiakim A, Mathieu PA. Reiners JJ Jr, et al. J Pharmacol Exp Ther. 2009 Mar;328(3):796-806. doi: 10.1124/jpet.108.144808. Epub 2008 Dec 9. J Pharmacol Exp Ther. 2009. PMID: 19066341 Free PMC article.
References
- Corbett T H, LoRusso P, Demchick L, Simpson C, Pugh S, White K, Kushner J, Polin L, Meyer J, Czarnecki J, et al. Invest New Drugs. 1998;16:129–139. - PubMed
- LoRusso P M, Demchick L, Knight J, Polin L, Behrens C H, McRipley R, Gross J L, Harrison B A, Corbett T H. Proc Am Assoc Cancer Res. 1994;35:401. (abstr.).
- Valeriote F, Corbett T, Edelstein M, Baker L. Clin Sci Rev. 1996;14:124–141. - PubMed
- Valeriote F, Moore R E, Patterson G M L, Paul V J, Scheurer P J, Corbett T. In: Anticancer Drug Discovery and Development: Natural Products and New Molecular Models. Valeriote F A, Corbett T H, Baker L H, editors. Boston: Kluwer; 1991. pp. 2–25.
- Subramanin B, Nakeff A, Valeriote F V. Proc Am Assoc Cancer Res. 1999;40:1. (abstr.).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials