DNA damage and replication checkpoints in fission yeast require nuclear exclusion of the Cdc25 phosphatase via 14-3-3 binding - PubMed (original) (raw)
DNA damage and replication checkpoints in fission yeast require nuclear exclusion of the Cdc25 phosphatase via 14-3-3 binding
Y Zeng et al. Mol Cell Biol. 1999 Nov.
Abstract
In fission yeast as well as in higher eukaryotic organisms, entry into mitosis is delayed in cells containing damaged or unreplicated DNA. This is accomplished in part by maintaining the Cdc25 phosphatase in a phosphorylated form that binds 14-3-3 proteins. In this study, we generated a mutant of fission yeast Cdc25 that is severely impaired in its ability to bind 14-3-3 proteins. Loss of both the DNA damage and replication checkpoints was observed in fission yeast cells expressing the 14-3-3 binding mutant. These findings indicate that 14-3-3 binding to Cdc25 is required for fission yeast cells to arrest their cell cycle in response to DNA damage and replication blocks. Furthermore, the 14-3-3 binding mutant localized almost exclusively to the nucleus, unlike wild-type Cdc25, which localized to both the cytoplasm and the nucleus. Nuclear accumulation of wild-type Cdc25 was observed when fission yeast cells were treated with leptomycin B, indicating that Cdc25 is actively exported from the nucleus. Nuclear exclusion of wild-type Cdc25 was observed upon overproduction of Rad 24, one of the two fission yeast 14-3-3 proteins, indicating that one function of Rad 24 is to keep Cdc25 out of the nucleus. In support of this conclusion, Rad 24 overproduction did not alter the nuclear location of the 14-3-3 binding mutant. These results indicate that 14-3-3 binding contributes to the nuclear exclusion of Cdc25 and that the nuclear exclusion of Cdc25 is required for a normal checkpoint response to both damaged and unreplicated DNA.
Figures
FIG. 1
Identification of 12 residues of Cdc25 phosphorylated by Cds1 in vitro. (A and B) Cdc25-WT and Cdc25-9A were purified as GST fusion proteins, and kinase assays were performed in vitro in the presence of the Cds1 protein kinase. Radiolabeled GST-Cdc25-WT (A) and GST-Cdc25-9A (B) were digested with trypsin, and the tryptic peptides were resolved by reverse-phase HPLC. Column fractions were collected and monitored for the presence of radioactivity. Identified residues are shown above the fraction in which they elute. Peptides containing phosphorylated S234 and S567/T569 elute in more than one fraction. (C) Phosphorylation sites (bold and italics) and neighboring residues. The consensus sequence for 14-3-3 binding as defined by Muslin et al. (40) is indicated.
FIG. 2
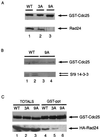
Association between Cdc25 and 14-3-3 proteins in vitro and in vivo. (A) Cdc25-WT (lane 1), Cdc25-3A (lane 2), and Cdc25-9A (lane 3) were purified as GST fusion proteins, and kinase assays were performed in vitro in the presence of the Cds1 protein kinase. Binding assays were then performed in the presence of purified Rad 24, and the reaction products were subjected to SDS-PAGE. Association of Rad 24 with Cdc25 was monitored by immunoblotting for Cdc25 (top) and Rad 24 (bottom). (B) Lysates were prepared from Sf9 insect cells infected with recombinant baculovirus encoding either GST-Cdc25-WT (WT; lanes 1 and 2) or GST-Cdc25-9A (lanes 3 and 4). GSH-agarose was used to precipitate the Cdc25 protein from either 400 μg (lanes 1 and 3) or 600 μg (lanes 2 and 4) of total cellular lysate. Precipitates were resolved by SDS- PAGE and immunoblotted for Cdc25 (top) and endogenous insect cell 14-3-3 proteins (bottom). (C) TE236 cells (leu1-32 ura4-D18 h−) cotransformed with pREP82-HA-rad24 and pREP3x-GST-cdc25(WT), pREP3x-GST-cdc25(3A), or pREP3x-GST-cdc25(9A) were grown in thiamine-free medium for ∼20 h. Lysates were prepared and analyzed directly by SDS-PAGE (lanes 1 to 3) or were incubated with GSH-agarose beads (lanes 4 to 6) prior to SDS-PAGE. Coprecipitation of Cdc25 with Rad 24 was analyzed by Western blotting. Lanes 1 to 3 represent 5% of the input lysate used in the precipitation assays. The experiments were all performed at least three times.
FIG. 3
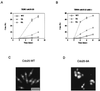
Mutation of canonical 14-3-3 binding sites disrupts the checkpoint response to unreplicated DNA. (A and B) cdc25-22 (TE282) (A) and _cdc25-22 mik1_− (TE938) (B) cells were transformed with pREP81-cdc25+ (WT), pREP81-cdc25(3A) (3A), or pREP81-cdc25(9A) (9A). Cells were treated with HU and analyzed as described previously (62). At the indicated times, cells were fixed and stained with DAPI and the percentage of cut cells was determined by counting at least 100 cells. Five to seven independent transformants were assayed for each data point. (C and D) Photographs of HU-treated cdc25+ _mik1_− cells (C) and _cdc25-9A mik1_− cells (D) at 6 h. Bar, 10 μm.
FIG. 4
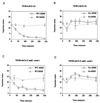
Mutation of canonical 14-3-3 binding sites disrupts the checkpoint response to damaged DNA. cdc25-22 (TE282) (A and B) and _cdc25-22 mik1_− (TE938) (C and D) cells were transformed with pREP81-cdc25+ (WT) or pREP81-cdc25(9A) (9A). Cells were either untreated (−MMS) or incubated with 0.1% MMS (+MMS). At the indicated times, samples were harvested and processed for Calcofluor and DAPI staining. The septation index [(number of septated cells/total number of cells) × 100] was determined. Three independent transformants were assayed for each data point.
FIG. 5
Localization of Cdc25 and phosphorylation site mutants in fission yeast. TE236 (_leu1-32 ura4-D18 h_−) transformed with pRep41x-GFP or pREP41x-GFP-cdc25(WT), pREP41x-GFP-cdc25(3A), or pREP41x-GFP-cdc25(9A), in the absence (A to H and M) or in the presence (I to L) of pREP2-HA-rad24 were grown in thiamine-free medium for ∼ 20 h. In some cases, cultures were incubated for 1 to 2 h with 20 ng of LMB per ml prior to analysis (E to H). In one case, cells were treated with 0.03% MMS and photographed 2 h later (M). Live cells were observed by using a conventional fluorescence microscope. Bar, 10 μm.
FIG. 6
Rad 24(I222A, L226A) is impaired in its ability to bind to Cdc25. (A) GST-Cdc25 was purified on GSH beads, phosphorylated by Cds1 in vitro, and incubated with ∼200 ng of Rad 24 or Rad 24(I222A, L226A). Precipitates were washed and subjected to SDS-PAGE. Association of Rad 24 with Cdc25 was monitored by immunoblotting for Cdc25 (top) and Rad 24 (bottom). Lanes: 1 and 2, 50 ng of Rad24 and Rad24(I222A, L226A) as input controls, respectively; 3 and 4, binding of phosphorylated GST-Cdc25 to Rad24 and Rad24(I222A, L226A), respectively. (B) TE236 cells (_leu1-32 ura4-D18 h_−) cotransformed with pREP3x-GST-cdc25(WT) and either pREP82-HA-rad24 (lanes 1 and 3) or pREP82-HA-rad24(I222A, L226A) (lanes 2 and 4) were induced for ∼ 20 h. Lysates were prepared and analyzed directly by SDS-PAGE (lanes 1 and 2) or were incubated with anti-HA antibody bound to protein A beads (lanes 3 and 4) prior to SDS-PAGE. Coimmunoprecipitation of Cdc25 with Rad 24 was analyzed by Western blotting. Lanes 1 and 2 represent 10% of the input lysate used in the precipitation assays. IgG, immunoglobulin G.
Similar articles
- Carboxy-terminal phosphorylation sites in Cdc25 contribute to enforcement of the DNA damage and replication checkpoints in fission yeast.
Frazer C, Young PG. Frazer C, et al. Curr Genet. 2012 Aug;58(4):217-34. doi: 10.1007/s00294-012-0379-1. Epub 2012 Jul 18. Curr Genet. 2012. PMID: 22806395 - Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1.
Zeng Y, Forbes KC, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Zeng Y, et al. Nature. 1998 Oct 1;395(6701):507-10. doi: 10.1038/26766. Nature. 1998. PMID: 9774107 - The fission yeast meiotic checkpoint kinase Mek1 regulates nuclear localization of Cdc25 by phosphorylation.
Pérez-Hidalgo L, Moreno S, San-Segundo PA. Pérez-Hidalgo L, et al. Cell Cycle. 2008 Dec;7(23):3720-30. doi: 10.4161/cc.7.23.7177. Epub 2008 Dec 13. Cell Cycle. 2008. PMID: 19029820 - Cdc25: mechanisms of checkpoint inhibition and recovery.
Karlsson-Rosenthal C, Millar JB. Karlsson-Rosenthal C, et al. Trends Cell Biol. 2006 Jun;16(6):285-92. doi: 10.1016/j.tcb.2006.04.002. Epub 2006 May 8. Trends Cell Biol. 2006. PMID: 16682204 Review. - Modelling the fission yeast cell cycle.
Sveiczer A, Tyson JJ, Novak B. Sveiczer A, et al. Brief Funct Genomic Proteomic. 2004 Feb;2(4):298-307. doi: 10.1093/bfgp/2.4.298. Brief Funct Genomic Proteomic. 2004. PMID: 15163365 Review.
Cited by
- The Integration of Proteome-Wide PTM Data with Protein Structural and Sequence Features Identifies Phosphorylations that Mediate 14-3-3 Interactions.
Egbert CM, Warr LR, Pennington KL, Thornton MM, Vaughan AJ, Ashworth SW, Heaton MJ, English N, Torres MP, Andersen JL. Egbert CM, et al. J Mol Biol. 2023 Jan 30;435(2):167890. doi: 10.1016/j.jmb.2022.167890. Epub 2022 Nov 17. J Mol Biol. 2023. PMID: 36402225 Free PMC article. - The Arabidopsis 14-3-3 protein, GF14omega, binds to the Schizosaccharomyces pombe Cdc25 phosphatase and rescues checkpoint defects in the rad24- mutant.
Sorrell DA, Marchbank AM, Chrimes DA, Dickinson JR, Rogers HJ, Francis D, Grierson CS, Halford NG. Sorrell DA, et al. Planta. 2003 Nov;218(1):50-7. doi: 10.1007/s00425-003-1083-7. Epub 2003 Aug 26. Planta. 2003. PMID: 12942327 - Redundant mechanisms prevent mitotic entry following replication arrest in the absence of Cdc25 hyper-phosphorylation in fission yeast.
Frazer C, Young PG. Frazer C, et al. PLoS One. 2011;6(6):e21348. doi: 10.1371/journal.pone.0021348. Epub 2011 Jun 23. PLoS One. 2011. PMID: 21731711 Free PMC article. - c-Myc alters the DNA damage-induced G2/M arrest in human mammary epithelial cells.
Sheen JH, Woo JK, Dickson RB. Sheen JH, et al. Br J Cancer. 2003 Oct 20;89(8):1479-85. doi: 10.1038/sj.bjc.6601307. Br J Cancer. 2003. PMID: 14562020 Free PMC article. - Carboxy-terminal phosphorylation sites in Cdc25 contribute to enforcement of the DNA damage and replication checkpoints in fission yeast.
Frazer C, Young PG. Frazer C, et al. Curr Genet. 2012 Aug;58(4):217-34. doi: 10.1007/s00294-012-0379-1. Epub 2012 Jul 18. Curr Genet. 2012. PMID: 22806395
References
- Alfa C E, Ducommun B, Beach D, Hyams J S. Distinct nuclear and spindle pole body populations of cyclin-cdc2 in fission yeast. Nature. 1990;347:680–682. - PubMed
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or the thiamine repressibility. Gene. 1993;123:131–136. - PubMed
- Blasina A, Van de Weyer I, Laus M C, Luyten W H M L, Parker A E, McGowan C H. A human homologue of the checkpoint kinase Cds1 directly inhibits Cdc25 phosphatase. Curr Biol. 1998;9:1–10. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials