The Rpb4 subunit of fission yeast Schizosaccharomyces pombe RNA polymerase II is essential for cell viability and similar in structure to the corresponding subunits of higher eukaryotes - PubMed (original) (raw)
The Rpb4 subunit of fission yeast Schizosaccharomyces pombe RNA polymerase II is essential for cell viability and similar in structure to the corresponding subunits of higher eukaryotes
H Sakurai et al. Mol Cell Biol. 1999 Nov.
Abstract
Both the gene and the cDNA encoding the Rpb4 subunit of RNA polymerase II were cloned from the fission yeast Schizosaccharomyces pombe. The cDNA sequence indicates that Rpb4 consists of 135 amino acid residues with a molecular weight of 15,362. As in the case of the corresponding subunits from higher eukaryotes such as humans and the plant Arabidopsis thaliana, Rpb4 is smaller than RPB4 from the budding yeast Saccharomyces cerevisiae and lacks several segments, which are present in the S. cerevisiae RPB4 subunit, including the highly charged sequence in the central portion. The RPB4 subunit of S. cerevisiae is not essential for normal cell growth but is required for cell viability under stress conditions. In contrast, S. pombe Rpb4 was found to be essential even under normal growth conditions. The fraction of RNA polymerase II containing RPB4 in exponentially growing cells of S. cerevisiae is about 20%, but S. pombe RNA polymerase II contains the stoichiometric amount of Rpb4 even at the exponential growth phase. In contrast to the RPB4 homologues from higher eukaryotes, however, S. pombe Rpb4 formed stable hybrid heterodimers with S. cerevisiae RPB7, suggesting that S. pombe Rpb4 is similar, in its structure and essential role in cell viability, to the corresponding subunits from higher eukaryotes. However, S. pombe Rpb4 is closer in certain molecular functions to S. cerevisiae RPB4 than the eukaryotic RPB4 homologues.
Figures
FIG. 1
Structure of the rpb4 gene and the Rpb4 protein. (A) Nucleotide sequences were determined for both the rpb4 gene and its cDNA. The amino acid sequence of Rpb4 was predicted from the cDNA sequence. The Rpb4 open reading frame (large black bars) is interrupted by three introns (small white bars). Nucleotide 1 is defined as the first nucleotide of the initiation codon, while amino acid 1 is defined as the initiation codon. The positions and directions of primers used for PCR are shown by the arrows. For primer sequences, see Table 1. (B) Comparison of the amino acid sequences of RNA polymerase II subunit 4 in various organisms. The amino acid sequence of S. pombe Rpb4 subunit (Sp) is compared with the corresponding subunits from S. cerevisiae (Sc), Homo sapiens (Hs), and A. thaliana (At). The overall identity of the amino acid (aa) sequence of the S. pombe Rpb4 with those of other organisms is shown at the end of each alignment. Amino acids that are identical or similar at least between two species are outlined or shaded, respectively. Gaps introduced to maximize alignment are indicated by dashes.
FIG. 2
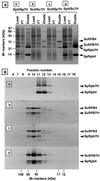
Identification of Rpb4 in purified S. pombe RNA polymerase II. (A) RNA polymerase II was purified by the standard procedure. The crude enzyme preparation after MonoQ chromatography was separated by SDS-PAGE and subjected to Western blotting with anti-Rpb7CH (lane 1), anti-Rpb8CH (lane 2), anti-Rpb9CH (lane 3), anti-Rpb4CH (lane 4), and anti-Rpb11CH (lane 5) antibodies. (B) Cell lysates of E. coli overexpressing the Rpb proteins (lanes 1 to 4, Rpb8, Rpb9, Rpb4, and Rpb11, respectively) were separated by SDS-PAGE, and the gel was subjected to either immunostaining against the anti-Rpb4 antibodies or protein staining with Coomassie brilliant blue (CBB). (C) The MonoQ fraction of RNA polymerase II was further purified by gel filtration chromatography on a Superose 6 column. Aliquots from Superose 6 fractions (lanes 1 to 10) and an aliquot of the Ni2+-affinity-purified RNA polymerase II from a S. pombe strain expressing His6-tagged Rpb3 (lane 11) were separated by SDS-PAGE. The gel was subjected to Western blotting with anti-Rpb4CH (top) and anti-GST-Rpb3 (bottom) antibodies. (D) The purified RNA polymerase II (Pol II) (Superose 6 fraction) was fractionated by SDS-PAGE (lane 4), and the gel was subjected to quantitative immunoblot analysis using anti-Rpb7CH (top) and anti-Rpb4CH antibodies (bottom). For quantitation, various amounts of the purified Rpb4 and Rpb7CH were analyzed in parallel (10 [lane 1], 20 [lane 2], and 50 [lane 3] ng).
FIG. 3
Tetrad analysis of an rpb4 heterozygous diploid. Diploid cells carrying one disrupted copy of rpb4 were sporulated on ME medium at 27°C for 2 days, and tetrads were dissected on YE medium containing adenine and uracil and allowed to grow at 30°C for 3 days. Seven tetrads are shown; the four spores from each tetrad are aligned vertically.
FIG. 4
Formation of complexes of Rpb4 or RPB4 and Rpb7 or RPB7. Two species of the RNA polymerase II subunit were coexpressed in E. coli in the following four combinations: S. pombe Rpb4-S. pombe Rpb7CH, S. cerevisiae RPB4-S. cerevisiae RPB7CH, S. cerevisiae RPB4-S. pombe Rpb7CH, and S. pombe Rpb4-S. cerevisiae RPB7CH, (which are shown as Sp4/Sp7H, Sc4/Sc7H, Sc4/Sp7H, and Sp4/Sc7H over the lanes in panel A). (A) Crude cell extracts were applied to a Ni2+-agarose column. Proteins bound were eluted with an elution buffer containing imidazole. Aliquots of the loading fraction, the flowthrough (FT) fraction, and the column-bound fraction were analyzed by SDS-PAGE, and the gel was stained with Coomassie brilliant blue. The migration positions of molecular mass markers are shown on the left. The positions of Rpb4, RPB4, Rpb7CH (Rpb7H), and RPB7CH (RPB7H) from S. pombe (Sp) or S. cerevisiae (Sc) are indicated on the right (note that the migration of S. pombe Rpb4 is faster than Rpb7). (B) The Ni2+-agarose elution fractions containing the binary complexes were subjected to gel filtration chromatography on a Superdex 75 PC 3.2/30 column. Aliquots from the fractions were analyzed by SDS-PAGE, and the gel was stained with Coomassie brilliant blue. Fraction numbers are shown at the top, and the peak positions of molecular marker proteins, fractionated on the same column, are indicated at the bottom. The migration positions of Rpb4, RPB4, Rpb7CH, and RPB7CH are indicated on the right.
Similar articles
- Rpb7 subunit of RNA polymerase II interacts with an RNA-binding protein involved in processing of transcripts.
Mitsuzawa H, Kanda E, Ishihama A. Mitsuzawa H, et al. Nucleic Acids Res. 2003 Aug 15;31(16):4696-701. doi: 10.1093/nar/gkg688. Nucleic Acids Res. 2003. PMID: 12907709 Free PMC article. - A comparative study of the proteome regulated by the Rpb4 and Rpb7 subunits of RNA polymerase II in fission yeast.
Kumar D, Varshney S, Sengupta S, Sharma N. Kumar D, et al. J Proteomics. 2019 May 15;199:77-88. doi: 10.1016/j.jprot.2019.03.007. Epub 2019 Mar 9. J Proteomics. 2019. PMID: 30862564 - Identification of the gene and the protein of RNA polymerase II subunit 9 (Rpb9) from the fission yeast Schizosacharomyces pombe.
Sakurai H, Kimura M, Ishihama A. Sakurai H, et al. Gene. 1998 Oct 9;221(1):11-6. doi: 10.1016/s0378-1119(98)00449-1. Gene. 1998. PMID: 9852944 - RNA polymerase II transcription apparatus in Schizosaccharomyces pombe.
Mitsuzawa H, Ishihama A. Mitsuzawa H, et al. Curr Genet. 2004 Jan;44(6):287-94. doi: 10.1007/s00294-003-0446-8. Epub 2003 Oct 22. Curr Genet. 2004. PMID: 14574615 Review. - Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe.
Huang Y. Huang Y. Nucleic Acids Res. 2002 Apr 1;30(7):1465-82. doi: 10.1093/nar/30.7.1465. Nucleic Acids Res. 2002. PMID: 11917007 Free PMC article. Review.
Cited by
- Multiple mechanisms of suppression circumvent transcription defects in an RNA polymerase mutant.
Tan Q, Li X, Sadhale PP, Miyao T, Woychik NA. Tan Q, et al. Mol Cell Biol. 2000 Nov;20(21):8124-33. doi: 10.1128/MCB.20.21.8124-8133.2000. Mol Cell Biol. 2000. PMID: 11027282 Free PMC article. - In vitro interaction between the N-terminus of the Ewing's sarcoma protein and the subunit of RNA polymerase II hsRPB7.
Todorova R. Todorova R. Mol Biol Rep. 2009 Jul;36(6):1269-74. doi: 10.1007/s11033-008-9308-2. Epub 2008 Jul 8. Mol Biol Rep. 2009. PMID: 18607770 - Rpb7 subunit of RNA polymerase II interacts with an RNA-binding protein involved in processing of transcripts.
Mitsuzawa H, Kanda E, Ishihama A. Mitsuzawa H, et al. Nucleic Acids Res. 2003 Aug 15;31(16):4696-701. doi: 10.1093/nar/gkg688. Nucleic Acids Res. 2003. PMID: 12907709 Free PMC article. - RNA polymerase II subunit D is essential for zebrafish development.
Maeta M, Kataoka M, Nishiya Y, Ogino K, Kashima M, Hirata H. Maeta M, et al. Sci Rep. 2020 Aug 6;10(1):13213. doi: 10.1038/s41598-020-70110-1. Sci Rep. 2020. PMID: 32764610 Free PMC article.
References
- Asturias F J, Meredith G D, Poglisch C L, Kornberg R D. Two conformations of RNA polymerase II revealed by electron crystallography. J Mol Biol. 1997;272:536–540. - PubMed
- Dezelee S, Francouise W, Sentenac A, Fromageot P. Two forms of RNA polymerase B in yeast. Eur J Biochem. 1976;65:543–552. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials