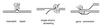Separation-of-function mutations in Saccharomyces cerevisiae MSH2 that confer mismatch repair defects but do not affect nonhomologous-tail removal during recombination - PubMed (original) (raw)
Separation-of-function mutations in Saccharomyces cerevisiae MSH2 that confer mismatch repair defects but do not affect nonhomologous-tail removal during recombination
B Studamire et al. Mol Cell Biol. 1999 Nov.
Abstract
Yeast Msh2p forms complexes with Msh3p and Msh6p to repair DNA mispairs that arise during DNA replication. In addition to their role in mismatch repair (MMR), the MSH2 and MSH3 gene products are required to remove 3' nonhomologous DNA tails during genetic recombination. The mismatch repair genes MSH6, MLH1, and PMS1, whose products interact with Msh2p, are not required in this process. We have identified mutations in MSH2 that do not disrupt genetic recombination but confer a strong defect in mismatch repair. Twenty-four msh2 mutations that conferred a dominant negative phenotype for mismatch repair were isolated. A subset of these mutations mapped to residues in Msh2p that were analogous to mutations identified in human nonpolyposis colorectal cancer msh2 kindreds. Approximately half of the these MMR-defective mutations retained wild-type or nearly wild-type activity for the removal of nonhomologous DNA tails during genetic recombination. The identification of mutations in MSH2 that disrupt mismatch repair without affecting recombination provides a first step in dissecting the Msh-effector protein complexes that are thought to play different roles during DNA repair and genetic recombination.
Figures
FIG. 1
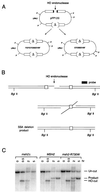
Map of mutated Msh2p residues in S. cerevisiae and in human HNPCC kindreds. Residues corresponding to the carboxyl termini of the two homologs are shown. Identical and similar residues are shaded black and gray, respectively. The dominant negative yeast mutations are indicated by solid arrows above the yeast Msh2 (yMsh2) sequence; mutations identified in the human kindreds are indicated by open arrows below the human Msh2 (hMsh2) sequence. Of the 59 human MSH2 mutations that were examined, 31 mapped to the region shown (see Materials and Methods). Yeast mutations in residues identical to HNPCC mutated residues are indicated by the boxes. Mutations in yeast MSH2 that confer wild-type activity in the removal of nonhomologous DNA during DSB repair are indicated by circles. The ATP binding domain is located between residues 686 and 700 (region I), 739 and 745 (region II), 762 and 773 (region III), and 798 and 803 (region IV) with respect to the yeast Msh2p sequence (14, 48). The location of the msh2-D924V mutation in the double-mutant allele p[_msh2-3_] is not shown. ∗, nonsense mutation.
FIG. 2
(A) Structure of pFP120 and predicted gene conversion events, with and without crossing over, following HO cleavage. P denotes _Pst_I sites that distinguish crossover from noncrossover recombinants. A detailed description of the structure of pFP120 can be found in reference . (B) SSA between identical 205-bp URA3 gene segments that flank an HO-induced DSB. Following HO cleavage, 5′-3′ exonuclease digestion produces 3′ single-stranded tails that can anneal with complementary sequences. Removal of 3′ nonhomologous tails results in the formation of the deletion product. _Bgl_II sites that distinguish uncut (7.4 kb) and HO-cut (4.8 kb) chromosomal DNA from the deletion product (5.5 kb) are indicated. (C) Southern blot analysis showing the kinetics of _HO_-induced SSA in wild-type (tNS1817 containing pJH727 and pEAA54), _msh2_Δ (tNS1817 containing pJH727), and msh2-R730W (tNS1817 containing pJH727 and pEAA83) strains that contain repeats of the 205-bp URA3 gene segment illustrated in panel B. Chromosomal DNA from the indicated strains was isolated at the indicated time points after HO induction, digested with _Bgl_II, and probed with a DNA fragment centromere proximal to the URA3 gene (66). The ratios of plasmid-bearing colonies after and before induction were 0.09, 1.8, and 1.2 for _msh2_Δ, MSH2, and msh2-R730W strains, respectively. These values reflect the value of product formation, the extent of induction, and the growth of the culture during the 5-h induction (66).
FIG. 3
Model describing Msh2p-Msh3p interactions with duplex DNA containing nucleotide insertion-deletion mismatches or 3′ tails. In this model, binding of Msh2p-Msh3p (–3) to a loop mismatch results in recruitment of the Mlh1p-Pms1p (L–M) complex and subsequent MMR steps. During SSA and gene conversion events, binding of Msh2p-Msh3p to duplex DNA containing 3′ tails results in stabilization of the recombination intermediate and allows the Rad1p-Rad10p (–10) endonuclease to cleave the 3′ tails. We propose that msh2 dominant negative mutations displaying a separation-of-function phenotype do not affect the ability of corresponding Msh2p-Msh3p complexes to bind to DNA mispairs or to duplex DNA containing 3′ tails. However, we hypothesize that these mutant complexes are specifically defective in interactions with downstream MMR factors such as Mlh1p-Pms1p but are capable of stabilizing recombination intermediates for the Rad1p-Rad10p endonuclease.
Similar articles
- Msh2 separation of function mutations confer defects in the initiation steps of mismatch repair.
Kijas AW, Studamire B, Alani E. Kijas AW, et al. J Mol Biol. 2003 Aug 1;331(1):123-38. doi: 10.1016/s0022-2836(03)00694-6. J Mol Biol. 2003. PMID: 12875840 - Novel dominant mutations in Saccharomyces cerevisiae MSH6.
Das Gupta R, Kolodner RD. Das Gupta R, et al. Nat Genet. 2000 Jan;24(1):53-6. doi: 10.1038/71684. Nat Genet. 2000. PMID: 10615127 - Saccharomyces cerevisiae MSH2-MSH3 and MSH2-MSH6 complexes display distinct requirements for DNA binding domain I in mismatch recognition.
Lee SD, Surtees JA, Alani E. Lee SD, et al. J Mol Biol. 2007 Feb 9;366(1):53-66. doi: 10.1016/j.jmb.2006.10.099. Epub 2006 Nov 3. J Mol Biol. 2007. PMID: 17157869 Free PMC article. - Mouse models for human DNA mismatch-repair gene defects.
Wei K, Kucherlapati R, Edelmann W. Wei K, et al. Trends Mol Med. 2002 Jul;8(7):346-53. doi: 10.1016/s1471-4914(02)02359-6. Trends Mol Med. 2002. PMID: 12114115 Review. - Roles of the DNA mismatch repair and nucleotide excision repair proteins during meiosis.
Kirkpatrick DT. Kirkpatrick DT. Cell Mol Life Sci. 1999 Mar;55(3):437-49. doi: 10.1007/s000180050300. Cell Mol Life Sci. 1999. PMID: 10228557 Free PMC article. Review.
Cited by
- Learning Yeast Genetics from Miro Radman.
Haber JE. Haber JE. Cells. 2021 Apr 20;10(4):945. doi: 10.3390/cells10040945. Cells. 2021. PMID: 33923882 Free PMC article. Review. - Coordination of Rad1-Rad10 interactions with Msh2-Msh3, Saw1 and RPA is essential for functional 3' non-homologous tail removal.
Eichmiller R, Medina-Rivera M, DeSanto R, Minca E, Kim C, Holland C, Seol JH, Schmit M, Oramus D, Smith J, Gallardo IF, Finkelstein IJ, Lee SE, Surtees JA. Eichmiller R, et al. Nucleic Acids Res. 2018 Jun 1;46(10):5075-5096. doi: 10.1093/nar/gky254. Nucleic Acids Res. 2018. PMID: 29660012 Free PMC article. - Rad51-mediated double-strand break repair and mismatch correction of divergent substrates.
Anand R, Beach A, Li K, Haber J. Anand R, et al. Nature. 2017 Apr 20;544(7650):377-380. doi: 10.1038/nature22046. Epub 2017 Apr 12. Nature. 2017. PMID: 28405019 Free PMC article. - EXO1 and MSH6 are high-copy suppressors of conditional mutations in the MSH2 mismatch repair gene of Saccharomyces cerevisiae.
Sokolsky T, Alani E. Sokolsky T, et al. Genetics. 2000 Jun;155(2):589-99. doi: 10.1093/genetics/155.2.589. Genetics. 2000. PMID: 10835383 Free PMC article. - Mutator dynamics in sexual and asexual experimental populations of yeast.
Raynes Y, Gazzara MR, Sniegowski PD. Raynes Y, et al. BMC Evol Biol. 2011 Jun 7;11:158. doi: 10.1186/1471-2148-11-158. BMC Evol Biol. 2011. PMID: 21649918 Free PMC article.
References
- Alani E, Chi N-W, Kolodner R D. The Saccharomyces cerevisiae Msh2 protein specifically binds to duplex oligonucleotides containing mismatched DNA base pairs and loop insertions. Genes Dev. 1995;9:234–247. - PubMed
- Alani E, Lee S, Kane M F, Griffith J, Kolodner R D. Saccharomyces cerevisiae MSH2, a mispaired base recognition protein, also recognizes Holliday junctions in DNA. J Mol Biol. 1997;265:289–301. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous