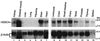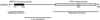HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor - PubMed (original) (raw)
HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor
A H Wang et al. Mol Cell Biol. 1999 Nov.
Abstract
Histone acetylation plays an important role in regulating chromatin structure and thus gene expression. Here we describe the functional characterization of HDAC4, a human histone deacetylase whose C-terminal part displays significant sequence similarity to the deacetylase domain of yeast HDA1. HDAC4 is expressed in various adult human tissues, and its gene is located at chromosome band 2q37. HDAC4 possesses histone deacetylase activity intrinsic to its C-terminal domain. When tethered to a promoter, HDAC4 represses transcription through two independent repression domains, with repression domain 1 consisting of the N-terminal 208 residues and repression domain 2 containing the deacetylase domain. Through a small region located at its N-terminal domain, HDAC4 interacts with the MADS-box transcription factor MEF2C. Furthermore, HDAC4 and MEF2C individually upregulate but together downmodulate c-jun promoter activity. These results suggest that HDAC4 interacts with transcription factors such as MEF2C to negatively regulate gene expression.
Figures
FIG. 1
Comparison of HDAC4-7 with HDA1. (A) Schematic representation of HDA1 and HDAC4 to -7. The N terminus of HDAC5 is incomplete, as are both termini of HDAC7. HDAC7N may be an alternatively spliced variant of HDAC7. The conserved deacetylase domains are boxed and labeled “DAC.” Other domains shared by HDAC4, -5, and -7 and HDAC7N are shown in bold lines. HDAC6 has a cysteine/histidine-rich domain (CH-rich; shaded box) at its C terminus. This diagram was generated based on BLAST search results. Sequences (GenBank accession numbers) referred to are HDA1 (P53973), HDAC4 (AB006626), HDAC5 (AB011172 and AF039691), HDAC6 (AJ011972), HDAC7 (AF124924), and HDAC7N (AB018287). A genomic clone (GenBank accession no. AC004466) contains some coding sequences related to HDAC4, -5, and -7 and may encode HDAC8. (B) Sequence alignment of catalytic domains of HDAC4 to -6 and HDA1. Identical or highly conserved residues (four of five sequences) are shaded. For simplicity, only S/T, R/K, and D/E are considered to be highly conserved. Asterisks denote histidines 802 and 803 of HDAC4, residues that may be important for deacetylase activity. (C) Sequence alignment of the N-terminal domains of HDAC4, -5, and -7N. Identical residues are shaded.
FIG. 2
Expression of HDAC4 in various adult human tissues. Poly(A) RNA blots (Clontech; 2 μg/lane) were probed with an HDAC4 cDNA fragment derived from the 3′ untranslated region (top). As a loading control, the same blots were reprobed with a β-actin cDNA probe (bottom). Molecular size markers are shown at the right.
FIG. 3
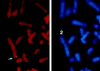
Chromosomal localization of the HDAC4 gene. Left, FISH signals detected at chromosome band 2q37.2, indicated by an arrow; right, the same mitotic cell stained with DAPI (4′,6-diamidino-2-phenylindole) to identify chromosomes. Human blood lymphocytes were used for FISH; the hybridization efficiency was 81% (i.e., 81 of 100 checked mitotic figures showed the indicated localization).
FIG. 4
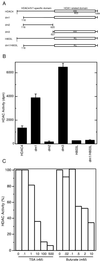
Characterization of histone deacetylase activity of HDAC4. (A) Schematic representation of HDAC4 and its mutants used for deacetylase assays. The letters “HH” denote histidines 802 and 803, which may be essential for deacetylase activity. (B) Deacetylase activity of HDAC4 and its mutants. Deacetylase activity, measured as disintegrations per minute of [3H]acetate released from [3H]acetyl-histones, was normalized to relative protein concentration determined by Western analyses with an anti-Flag antibody. During purification of Flag-tagged proteins, a buffer containing 0.5 M KCl was used for extensive washing; under such conditions, with untransfected cell extracts, equivalent amounts of M2-agarose beads retained deacetylase activity close to background levels. (C) Effects of TSA and sodium butyrate on deacetylase activity of dm3.
FIG. 5
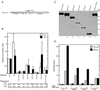
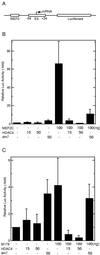
Tethered HDAC4 represses transcription. (A) Schematic representation of the luciferase reporter Gal4-tk-Luc. Upstream from the tk core promoter (−152 to +50) are five copies of the Gal4-binding site. (B) Repression of Gal4-tk-Luc by HDAC4 and its mutants in NIH 3T3 cells. The mutants dm1 to -3 and dm1/H803A are depicted in Fig. 4A; dm4 and dm5 contain the N-terminal 208 and 114 residues of HDAC4, respectively. Mammalian constructs expressing HDAC4 and its mutants fused to the C terminus of Gal4(1-147) were transfected into NIH 3T3 cells with the reporter Gal4-tk-Luc. Luciferase (Luc) activities were normalized to the internal β-galactosidase control; the normalized luciferase activity from the transfection without any effector plasmid was arbitrarily set to 1.0. (C) Expression of Gal4-HDAC4 and its mutants. Extracts (10 μg/lane), prepared from 293T cells transfected with expression plasmids for indicated fusion proteins, were subjected to Western blotting analyses using an anti-Gal4 antibody (RK5C1; Santa Cruz Biotechnology). Molecular size markers are shown at the right. (D) Repression of reporters with different core promoters by Gal4-dm3 in 293T cells. The reporters possess indicated core promoters replacing the tk region of Gal4-tk-Luc (A); 100 and 300 ng of expression plasmids were used as indicated.
FIG. 6
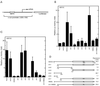
HDAC4 interacts with MEF2 in vivo and in vitro. (A) Immunoprecipitation of HDAC4 with MEF2C (lanes 1 to 5) or MEF2D (lanes 6 to 9). Flag-tagged HDAC4 (lanes 1 to 4 and 7) or dm4 (lanes 5 and 9) was expressed with (lanes 2, 4, and 5) or without (lanes 1, 3, and 6 to 9) MEF2C in 293T cells and immunoprecipitated (IP) with anti-Flag M2-agarose. Extracts (lanes 1, 2, and 6) and immunoprecipitated proteins eluted with Flag peptide (lanes 3 to 5 and 7 to 9) were subjected to Western blotting analyses with an anti-MEF2C (lanes 1 to 5) or anti-MEF2D (lanes 6 to 9) polyclonal antibody. The presence of Flag-tagged HDAC4 and dm4 was confirmed by Western blotting analyses of the same samples with an anti-Flag monoclonal antibody (data not shown). (B) Schematic representation of MEF2C and its mutant M178 (consisting residues 1 to 178). (C and D) Interaction of M178 with HDAC4 and its deletion mutants in vitro. MBP or MBP-M178 was immobilized on amylose-agarose and tested for interaction with HDAC4 or its deletion mutants, synthesized in vitro in the presence of [35S]methionine. Input lanes represent 20% of HDAC4 or its mutants used for interaction. (E) Schematic representation of HDAC4 and its deletion mutants used in the interaction assays (A, C, and D). The + symbol denotes that the protein shown at left interacts with MEF2C.
FIG. 7
HDAC4 represses transcription in a MEF2C-dependent manner. (A) Schematic representation of the reporter MEF2-E4-Luc, which contains one copy of the MEF2-binding site upstream from the adenovirus E4 core promoter (−34 to +34) and the luciferase coding sequence. (B) HDAC4 represses MEF2C-dependent transcription. MEF2-E4-Luc was cotransfected into NIH 3T3 cells with the expression plasmids at indicated amounts. Luciferase (Luc) activities were normalized to the internal β-galactosidase control; the normalized luciferase activity from the transfection without any effector plasmid was arbitrarily set to 1.0. (C) Recruitment of HDAC4 by the MEF2C mutant M178 leads to repression. Reporter assays were performed as for panel B except that the expression plasmid for M178 was used instead.
FIG. 8
HDAC4 and MEF2C cooperatively regulate c-jun promoter activity. (A) Schematic representation of the reporter pJLuc, which contains positions −225 to +150 of the c-jun promoter upstream of the luciferase coding sequence. (B) HDAC4 activates c-jun promoter activity in a MEF2C-independent manner. The pJLuc reporter and expression plasmids for HDAC4 or its mutants were cotransfected into NIH 3T3 cells. Luciferase (Luc) activities were normalized to the internal β-galactosidase control; the normalized luciferase activity from the transfection without any effector plasmid was arbitrarily set to 1.0. (C) HDAC4 represses MEF2C-dependent transcription. Together with the MEF2C expression plasmid, pJLuc and indicated HDAC4 plasmids were cotransfected into NIH 3T3 cells. Reporter assays were determined and calculated as for panel B. (D) Schematic representation of HDAC4 and its mutants used for panels B and C. Repression of MEF2C-dependent transcription by each construct is shown at the right.
FIG. 9
Functional domain organization of HDAC4. HDAC4 possesses at least two repression domains, with repression domain 1 located at the N terminus and repression domain 2 at the C-terminal part including the HDA1-related deacetylase domain. The MEF2C interaction domain resides within the N-terminal domain; the region C-terminal from the MEF2C interaction domain may be involved in activation of the c-jun promoter in the absence of transfected MEF2C.
Similar articles
- HDAC4 deacetylase associates with and represses the MEF2 transcription factor.
Miska EA, Karlsson C, Langley E, Nielsen SJ, Pines J, Kouzarides T. Miska EA, et al. EMBO J. 1999 Sep 15;18(18):5099-107. doi: 10.1093/emboj/18.18.5099. EMBO J. 1999. PMID: 10487761 Free PMC article. - Histone deacetylase 4 possesses intrinsic nuclear import and export signals.
Wang AH, Yang XJ. Wang AH, et al. Mol Cell Biol. 2001 Sep;21(17):5992-6005. doi: 10.1128/MCB.21.17.5992-6005.2001. Mol Cell Biol. 2001. PMID: 11486037 Free PMC article. - mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity.
Lemercier C, Verdel A, Galloo B, Curtet S, Brocard MP, Khochbin S. Lemercier C, et al. J Biol Chem. 2000 May 19;275(20):15594-9. doi: 10.1074/jbc.M908437199. J Biol Chem. 2000. PMID: 10748098 - Class II histone deacetylases: structure, function, and regulation.
Bertos NR, Wang AH, Yang XJ. Bertos NR, et al. Biochem Cell Biol. 2001;79(3):243-52. Biochem Cell Biol. 2001. PMID: 11467738 Review. - Beside the MEF2 axis: unconventional functions of HDAC4.
Clocchiatti A, Di Giorgio E, Demarchi F, Brancolini C. Clocchiatti A, et al. Cell Signal. 2013 Jan;25(1):269-76. doi: 10.1016/j.cellsig.2012.10.002. Epub 2012 Oct 11. Cell Signal. 2013. PMID: 23063464 Review.
Cited by
- Strategies to minimize hypertrophy in cartilage engineering and regeneration.
Chen S, Fu P, Cong R, Wu H, Pei M. Chen S, et al. Genes Dis. 2015 Mar 1;2(1):76-95. doi: 10.1016/j.gendis.2014.12.003. Genes Dis. 2015. PMID: 26000333 Free PMC article. - Acetyl-lysine erasers and readers in the control of pulmonary hypertension and right ventricular hypertrophy.
Stratton MS, McKinsey TA. Stratton MS, et al. Biochem Cell Biol. 2015 Apr;93(2):149-57. doi: 10.1139/bcb-2014-0119. Epub 2014 Dec 16. Biochem Cell Biol. 2015. PMID: 25707943 Free PMC article. Review. - Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors.
Grégoire S, Yang XJ. Grégoire S, et al. Mol Cell Biol. 2005 Mar;25(6):2273-87. doi: 10.1128/MCB.25.6.2273-2287.2005. Mol Cell Biol. 2005. PMID: 15743823 Free PMC article. - Loss of the putative catalytic domain of HDAC4 leads to reduced thermal nociception and seizures while allowing normal bone development.
Rajan I, Savelieva KV, Ye GL, Wang CY, Malbari MM, Friddle C, Lanthorn TH, Zhang W. Rajan I, et al. PLoS One. 2009 Aug 12;4(8):e6612. doi: 10.1371/journal.pone.0006612. PLoS One. 2009. PMID: 19672313 Free PMC article. - The Therapeutic Strategy of HDAC6 Inhibitors in Lymphoproliferative Disease.
Cosenza M, Pozzi S. Cosenza M, et al. Int J Mol Sci. 2018 Aug 9;19(8):2337. doi: 10.3390/ijms19082337. Int J Mol Sci. 2018. PMID: 30096875 Free PMC article. Review.
References
- Ausio J, van Holde K E. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry. 1986;25:1421–1428. - PubMed
- Bartholomew C, Kilbey A, Clark A M, Walker M. The Evi-1 proto-oncogene encodes a transcriptional repressor activity associated with transformation. Oncogene. 1997;14:569–577. - PubMed
- Black B L, Olson E N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. - PubMed
- Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous