Calibrating bacterial evolution - PubMed (original) (raw)
Calibrating bacterial evolution
H Ochman et al. Proc Natl Acad Sci U S A. 1999.
Abstract
Attempts to calibrate bacterial evolution have relied on the assumption that rates of molecular sequence divergence in bacteria are similar to those of higher eukaryotes, or to those of the few bacterial taxa for which ancestors can be reliably dated from ecological or geological evidence. Despite similarities in the substitution rates estimated for some lineages, comparisons of the relative rates of evolution at different classes of nucleotide sites indicate no basis for their universal application to all bacteria. However, there is evidence that bacteria have a constant genome-wide mutation rate on an evolutionary time scale but that this rate differs dramatically from the rate estimated by experimental methods.
Figures
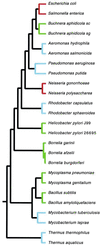
Figure 1
Phylogenetic tree of bacterial taxa used in this study based on 16S rRNA sequences. Only terminal branch lengths are proportional to sequence divergence. Colors indicate chromosomal G+C content as follows: green, <45% G+C; red, 45–55% G+C; blue, >55% G+C.
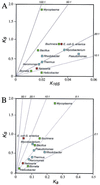
Figure 2
Relative extent of sequence divergence among pairs of bacterial species. Only pairs of species that differ by less than 5% in 16S rRNA sequences and for which sequence information is available for at least five homologous genes are included. Colors correspond to chromosomal G+C contents as in Fig. 1. (A) Divergence at synonymous sites (_K_s) vs. divergence at 16S rRNA (_K_16S). (B) Divergence at synonymous (_K_s) vs. nonsynonymous (_K_a) sites.
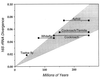
Figure 3
Rates of 16S rRNA sequence divergence in bacterial endosymbionts of insects. Divergence times are estimated from fossil evidence of insect hosts and were obtained from the literature (, –36). Shaded area denotes a rate of 16S rRNA divergence of 1–2% per 50 million years. Insect and bacterial taxa are follows (host/symbiont): aphids (Schizaphis graminum/Buchnera aphidicola, Schlechtendalia chinesis/Buchnera aphidicola); cockroach-termite (Blattaria germanica/symbiont, Mastotermes darwiniensis/symbiont); cockroach (Blattaria germanica/symbiont_, Periplaneta americana_/symbiont); whitefly (Trialeurodes vaporariorum/P-endosymbiont, Siphoninus phillyreae/P-endosymbiont); tsetse fly (Glossinia brevipalis/Wigglesworthia glossinidia, Glossinia morsitans/Wigglesworthia glossinidia).
Similar articles
- Inferring clocks when lacking rocks: the variable rates of molecular evolution in bacteria.
Kuo CH, Ochman H. Kuo CH, et al. Biol Direct. 2009 Sep 29;4:35. doi: 10.1186/1745-6150-4-35. Biol Direct. 2009. PMID: 19788732 Free PMC article. - The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification.
Cavalier-Smith T. Cavalier-Smith T. Int J Syst Evol Microbiol. 2002 Jan;52(Pt 1):7-76. doi: 10.1099/00207713-52-1-7. Int J Syst Evol Microbiol. 2002. PMID: 11837318 Review. - Genome-scale rates of evolutionary change in bacteria.
Duchêne S, Holt KE, Weill FX, Le Hello S, Hawkey J, Edwards DJ, Fourment M, Holmes EC. Duchêne S, et al. Microb Genom. 2016 Nov 30;2(11):e000094. doi: 10.1099/mgen.0.000094. eCollection 2016 Nov. Microb Genom. 2016. PMID: 28348834 Free PMC article. - Evolution in bacteria: evidence for a universal substitution rate in cellular genomes.
Ochman H, Wilson AC. Ochman H, et al. J Mol Evol. 1987;26(1-2):74-86. doi: 10.1007/BF02111283. J Mol Evol. 1987. PMID: 3125340 - DNA methylation: evolution of a bacterial immune function into a regulator of gene expression and genome structure in higher eukaryotes.
Bestor TH. Bestor TH. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1235):179-87. doi: 10.1098/rstb.1990.0002. Philos Trans R Soc Lond B Biol Sci. 1990. PMID: 1968655 Review.
Cited by
- Adaptive differentiation and rapid evolution of a soil bacterium along a climate gradient.
Chase AB, Weihe C, Martiny JBH. Chase AB, et al. Proc Natl Acad Sci U S A. 2021 May 4;118(18):e2101254118. doi: 10.1073/pnas.2101254118. Proc Natl Acad Sci U S A. 2021. PMID: 33906949 Free PMC article. - Population expansions shared among coexisting bacterial lineages are revealed by genetic evidence.
Avitia M, Escalante AE, Rebollar EA, Moreno-Letelier A, Eguiarte LE, Souza V. Avitia M, et al. PeerJ. 2014 Dec 16;2:e696. doi: 10.7717/peerj.696. eCollection 2014. PeerJ. 2014. PMID: 25548732 Free PMC article. - Phylogenetic tree construction using trinucleotide usage profile (TUP).
Chen S, Deng LY, Bowman D, Shiau JH, Wong TY, Madahian B, Lu HH. Chen S, et al. BMC Bioinformatics. 2016 Oct 6;17(Suppl 13):381. doi: 10.1186/s12859-016-1222-3. BMC Bioinformatics. 2016. PMID: 27766939 Free PMC article. - Secondary (gamma-Proteobacteria) endosymbionts infect the primary (beta-Proteobacteria) endosymbionts of mealybugs multiple times and coevolve with their hosts.
Thao ML, Gullan PJ, Baumann P. Thao ML, et al. Appl Environ Microbiol. 2002 Jul;68(7):3190-7. doi: 10.1128/AEM.68.7.3190-3197.2002. Appl Environ Microbiol. 2002. PMID: 12088994 Free PMC article. - Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis.
Gutierrez MC, Brisse S, Brosch R, Fabre M, Omaïs B, Marmiesse M, Supply P, Vincent V. Gutierrez MC, et al. PLoS Pathog. 2005 Sep;1(1):e5. doi: 10.1371/journal.ppat.0010005. Epub 2005 Aug 19. PLoS Pathog. 2005. PMID: 16201017 Free PMC article.
References
- Ochman H, Wilson A C. In: Escherichia coli and Salmonella typhimurium: Molecular and Cellular Aspects, Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1987. pp. 1649–1654.
- Ochman H, Wilson A C. J Mol Evol. 1987;26:74–86. - PubMed
- Moran N A, Munson M A, Baumann P, Ishikawa H. Proc R Soc London Ser B. 1993;253:167–171.
- Wernegreen J J, Moran N A. Mol Biol Evol. 1999;16:83–97. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources