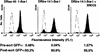XRCC3 promotes homology-directed repair of DNA damage in mammalian cells - PubMed (original) (raw)
XRCC3 promotes homology-directed repair of DNA damage in mammalian cells
A J Pierce et al. Genes Dev. 1999.
Abstract
Homology-directed repair of DNA damage has recently emerged as a major mechanism for the maintenance of genomic integrity in mammalian cells. The highly conserved strand transferase, Rad51, is expected to be critical for this process. XRCC3 possesses a limited sequence similarity to Rad51 and interacts with it. Using a novel fluorescence-based assay, we demonstrate here that error-free homology-directed repair of DNA double-strand breaks is decreased 25-fold in an XRCC3-deficient hamster cell line and can be restored to wild-type levels through XRCC3 expression. These results establish that XRCC3-mediated homologous recombination can reverse DNA damage that would otherwise be mutagenic or lethal.
Figures
Figure 1
GFP expression plasmids. (A) Modified GFP gene. The modified GFP gene encodes the EGFP protein fused to a nuclear localization signal (N) and zinc finger domain (Z). It is expressed from a hCMV enhancer/chicken β-actin promoter (arrow) on a spliced message (not shown). GFP is modified to SceGFP so as to contain an I-_Sce_I site and in-frame termination codons (underline). (B) DR–GFP recombination substrate. Downstream of the SceGFP gene is iGFP, a 5′ and 3′-truncated GFP gene. (C) (STGC) product. In a STGC, a DSB at the I-Sce I site is repaired from the iGFP gene on the same chromatid or sister chromatid, to result in a functional GFP gene. (D) Homology-mediated deletion product.
Figure 2
The irs1SF cell line is defective for DSB-induced homologous recombination because of XRCC3 deficiency. Wild-type (DRaa) and XRCC3-deficient (DRirs) cell lines were electroporated with each of the indicated expression vectors (100 μg of the I-Sce I vector; 20 μg of the XRCC3 vector), and 5 × 104 cells were analyzed by flow cytometry. (A) Two-color fluorescence analysis for cell lines DRaa-40 and DRirs-10. The percentage of green fluorescent cells falling above the diagonal for each transfection are indicated. (FL1) Green fluorescence; (FL2) orange fluorescence. (B) Summary of results from several cell lines.
Figure 3
Green fluorescence is stably expressed after I-Sce I expression. Wild-type DRaa-40 cells and XRCC3-deficient DRirs-14 cells were electroporated with the I-Sce I expression vector or both the I-Sce I and XRCC3 expression vectors, sorted on the basis of green fluorescence, and then reanalyzed by flow cytometry. Percent GFP+ cells before sorting (unshaded peak) and after sorting (shaded peaks) are indicated. Not shown is the DRaa-40 cell line transfected with both the I-Sce I and XRCC3 expression vectors, in which the presorted population was 0.66% GFP+ and the post-sorted population was 93.5% GFP+. Also not shown are the _lacZ_-transfected cells that were not sorted; 0.01% GFP+ for DRaa-40 and <0.01% GFP+ for DRirs-14. (The frequency of GFP+ cells is lower in this experiment than the one shown in Fig. 2, because only 20 μg of each of the expression vectors was electroporated.)
Figure 4
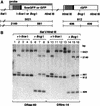
Green fluorescence is a measure of STGC. (A) Map of the parental or recombined DR–GFP substrate containing SceGFP or GFP, respectively. Sizes of expected fragments are indicated in base pairs. (B) Southern blot analysis of sorted cell lines. Genomic DNA was digested with the indicated endonucleases. (Lanes 1,2,5,6) Untransfected wild-type cell line DRaa-40; (lanes 3,7) GFP+ DRaa-40 cells from transfection of the I-Sce I expression vector; (lanes 4,8) GFP+ DRaa-40 cells from cotransfection of the I-Sce I and XRCC3 expression vectors; (lanes 9,10,13,14) untransfected mutant cell line DRirs-14; (lanes 11,15) GFP+ DRirs-14 cells from transfection of the I-Sce I expression vector; (lanes 12,16) GFP+ DRirs-14 cells from cotransfection of the I-Sce I and XRCC3 expression vectors. The residual 3021-bp band after _Bcg_I digestion in lanes 7,8,15, and 16 is likely due to GFP-negative cells in the population (see Fig. 3).
Figure 5
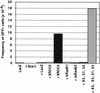
DSB-induced recombination is restored in the irs1SF cell line by transient expression of XRCC3 but not other recombination proteins. The DRirs-14 cell line was electroporated with each of the indicated expression vectors (20 μg). The total amount of DNA electroporated was maintained at a constant level by the addition of pCMV–lacZ. A total of 5 × 105 cells were analyzed by flow cytometry. (hRad51 or 51) Human Rad51; (mRad52 or 52) mouse Rad52; (X2) XRCC2; (X3) XRCC3.
Similar articles
- XRCC3 is required for efficient repair of chromosome breaks by homologous recombination.
Brenneman MA, Weiss AE, Nickoloff JA, Chen DJ. Brenneman MA, et al. Mutat Res. 2000 Mar 20;459(2):89-97. doi: 10.1016/s0921-8777(00)00002-1. Mutat Res. 2000. PMID: 10725659 - Xrcc3 is recruited to DNA double strand breaks early and independent of Rad51.
Forget AL, Bennett BT, Knight KL. Forget AL, et al. J Cell Biochem. 2004 Oct 15;93(3):429-36. doi: 10.1002/jcb.20232. J Cell Biochem. 2004. PMID: 15372620 - XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination.
Brenneman MA, Wagener BM, Miller CA, Allen C, Nickoloff JA. Brenneman MA, et al. Mol Cell. 2002 Aug;10(2):387-95. doi: 10.1016/s1097-2765(02)00595-6. Mol Cell. 2002. PMID: 12191483 - The role of homologous recombination processes in the repair of severe forms of DNA damage in mammalian cells.
Thacker J. Thacker J. Biochimie. 1999 Jan-Feb;81(1-2):77-85. doi: 10.1016/s0300-9084(99)80041-8. Biochimie. 1999. PMID: 10214913 Review. - The role of homologous recombination repair in the formation of chromosome aberrations.
Griffin CS, Thacker J. Griffin CS, et al. Cytogenet Genome Res. 2004;104(1-4):21-7. doi: 10.1159/000077462. Cytogenet Genome Res. 2004. PMID: 15162011 Review.
Cited by
- CBP and p300 histone acetyltransferases contribute to homologous recombination by transcriptionally activating the BRCA1 and RAD51 genes.
Ogiwara H, Kohno T. Ogiwara H, et al. PLoS One. 2012;7(12):e52810. doi: 10.1371/journal.pone.0052810. Epub 2012 Dec 20. PLoS One. 2012. PMID: 23285190 Free PMC article. - Targeting acute myeloid leukemia dependency on VCP-mediated DNA repair through a selective second-generation small-molecule inhibitor.
Roux B, Vaganay C, Vargas JD, Alexe G, Benaksas C, Pardieu B, Fenouille N, Ellegast JM, Malolepsza E, Ling F, Sodaro G, Ross L, Pikman Y, Conway AS, Tang Y, Wu T, Anderson DJ, Le Moigne R, Zhou HJ, Luciano F, Hartigan CR, Galinsky I, DeAngelo DJ, Stone RM, Auberger P, Schenone M, Carr SA, Guirouilh-Barbat J, Lopez B, Khaled M, Lage K, Hermine O, Hemann MT, Puissant A, Stegmaier K, Benajiba L. Roux B, et al. Sci Transl Med. 2021 Mar 31;13(587):eabg1168. doi: 10.1126/scitranslmed.abg1168. Sci Transl Med. 2021. PMID: 33790022 Free PMC article. - Heavy water inhibits DNA double-strand break repairs and disturbs cellular transcription, presumably via quantum-level mechanisms of kinetic isotope effects on hydrolytic enzyme reactions.
Yasuda T, Nakajima N, Ogi T, Yanaka T, Tanaka I, Gotoh T, Kagawa W, Sugasawa K, Tajima K. Yasuda T, et al. PLoS One. 2024 Oct 3;19(10):e0309689. doi: 10.1371/journal.pone.0309689. eCollection 2024. PLoS One. 2024. PMID: 39361575 Free PMC article. - Targeting cancer with a lupus autoantibody.
Hansen JE, Chan G, Liu Y, Hegan DC, Dalal S, Dray E, Kwon Y, Xu Y, Xu X, Peterson-Roth E, Geiger E, Liu Y, Gera J, Sweasy JB, Sung P, Rockwell S, Nishimura RN, Weisbart RH, Glazer PM. Hansen JE, et al. Sci Transl Med. 2012 Oct 24;4(157):157ra142. doi: 10.1126/scitranslmed.3004385. Sci Transl Med. 2012. PMID: 23100628 Free PMC article. - MCM8-9 complex promotes resection of double-strand break ends by MRE11-RAD50-NBS1 complex.
Lee KY, Im JS, Shibata E, Park J, Handa N, Kowalczykowski SC, Dutta A. Lee KY, et al. Nat Commun. 2015 Jul 28;6:7744. doi: 10.1038/ncomms8744. Nat Commun. 2015. PMID: 26215093 Free PMC article.
References
- Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23:247–251. - PubMed
- Bishop DK, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum RR, Shinohara A. Xrcc3 is required for assembly of Rad51 complexes in vivo. J Biol Chem. 1998;273:21482–21488. - PubMed
- Caldecott K, Jeggo P. Cross-sensitivity of gamma-ray-sensitive hamster mutants to cross-linking agents. Mutat Res. 1991;255:111–121. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials