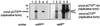The Vibrio cholerae O139 Calcutta bacteriophage CTXphi is infectious and encodes a novel repressor - PubMed (original) (raw)
The Vibrio cholerae O139 Calcutta bacteriophage CTXphi is infectious and encodes a novel repressor
B M Davis et al. J Bacteriol. 1999 Nov.
Abstract
CTXphi is a lysogenic, filamentous bacteriophage. Its genome includes the genes encoding cholera toxin (ctxAB), one of the principal virulence factors of Vibrio cholerae; consequently, nonpathogenic strains of V. cholerae can be converted into toxigenic strains by CTXphi infection. O139 Calcutta strains of V. cholerae, which were linked to cholera outbreaks in Calcutta, India, in 1996, are novel pathogenic strains that carry two distinct CTX prophages integrated in tandem: CTX(ET), the prophage previously characterized within El Tor strains, and a new CTX Calcutta prophage (CTX(calc)). We found that the CTX(calc) prophage gives rise to infectious virions; thus, CTX(ET)phi is no longer the only known vector for transmission of ctxAB. The most functionally significant differences between the nucleotide sequences of CTX(calc)phi and CTX(ET)phi are located within the phages' repressor genes (rstR(calc) and rstR(ET), respectively) and their RstR operators. RstR(calc) is a novel, allele-specific repressor that regulates replication of CTX(calc)phi by inhibiting the activity of the rstA(calc) promoter. RstR(calc) has no inhibitory effect upon the classical and El Tor rstA promoters, which are instead regulated by their cognate RstRs. Consequently, production of RstR(calc) renders a CTX(calc) lysogen immune to superinfection by CTX(calc)phi but susceptible (heteroimmune) to infection by CTX(ET)phi. Analysis of the prophage arrays generated by sequentially integrated CTX phages revealed that pathogenic V. cholerae O139 Calcutta probably arose via infection of an O139 CTX(ET)phi lysogen by CTX(calc)phi.
Figures
FIG. 1
Structure and sequence of CTX prophages within AS207, an O139 Calcutta strain of V. cholerae. (A) Within the AS207 chromosome, an RS1 element and a CTXET prophage are followed by two CTXcalc prophages (shown in light grey). The Calcutta prophage is structurally similar to the previously described CTX prophages within El Tor and classical strains, which contain two major domains known as RS2 and core. RS2 contains three genes (rstA, rstB, and rstR) whose transcriptional orientation is indicated by the arrows. The black triangles represent _attRS_-ER sequences. (B) Alignment of the nucleotide sequences of rstR, ig-2, and parts of rstA and ig-1 from Calcutta, El Tor, and classical prophages. The arrows underneath the sequences depict the ORFs that encode the three variants of RstR.
FIG. 2
Detection of transfer of CTXcalcφ and CTXETφ from supernatants of AS207 to O395 using Southern blot analysis of plasmid DNA. Undigested plasmid DNAs were run on agarose gels, transferred to a nylon membrane, and sequentially hybridized with probes for _rstR_calc (left panel) and _rstR_ET (right panel). Plasmid DNA was prepared from O395 (lanes 1), AS207 (lanes 2), 2740-80 (CTXET-Kn) (lanes 3), O395 cultured with AS207 supernatant (lanes 4), and O395 cultured with 2740-80 (CTXET-Kn) supernatant (lanes 5). The Knr cassette is slightly larger than the ctxAB genes it replaces in CTXET-Knφ, so pCTXET-Knφ migrates more slowly than pCTXETφ.
FIG. 3
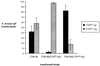
Relative efficiency of transformation of pCTXcalc-Ap versus pCTXET-Ap into 2740-80, 2740-80 (CTXET-Kn), and 2740-80 (CTXcalc-Kn). Percentage of total Apr transformants = (pCTXcalcAp or pCTXETAp transformants)/(pCTXcalcAp + pCTXETAp transformants).
FIG. 4
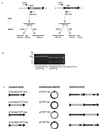
Integration site selection by sequentially integrated plasmids. (A) PCR followed by restriction digestion was used to determine the order of prophages within the chromosome following sequential integration of CTXET and CTXcalc antibiotic-marked plasmids. When the CTXcalc prophage is the furthest 5′, the TLCF1-RstR Rev PCR product contains a _Hin_dIII site but not a _Bgl_II site. Conversely, when CTXET is upstream, the PCR product contains a _Bgl_II site but not a _Hin_dIII site. (B) A representative agarose gel containing PCR products digested with _Hin_dIII. Template DNA is as follows: 2740-80(CTXET-Kn) transformed with pCTXcalc-Ap (lanes 1 to 6), 2740-80 (CTXcalc-Kn) transformed with pCTXET-Ap (lanes 7 to 12), and 2740-80 (CTXcalc-Kn) (lane 13). (C) Summary and model of integration site selection following sequential CTXφ integration. Phage DNA does not integrate into the 5′ ER (black triangle) if the chromosome already contains a CTX prophage. Instead, the new phage DNA inserts into the 3′ ER.
Similar articles
- A novel pre-CTX prophage in the Vibrio cholerae serogroup O139 strain.
Li X, Zhao L, Gao H, Chen L, Fan F, Li Z, Fan Y, Li J, Liang W, Pang B, Kan B. Li X, et al. Infect Genet Evol. 2020 Jul;81:104238. doi: 10.1016/j.meegid.2020.104238. Epub 2020 Feb 8. Infect Genet Evol. 2020. PMID: 32045711 - Genesis of variants of Vibrio cholerae O1 biotype El Tor: role of the CTXphi array and its position in the genome.
Nandi S, Maiti D, Saha A, Bhadra RK. Nandi S, et al. Microbiology (Reading). 2003 Jan;149(Pt 1):89-97. doi: 10.1099/mic.0.25599-0. Microbiology (Reading). 2003. PMID: 12576583 - Molecular analysis of the rstR and orfU genes of the CTX prophages integrated in the small chromosomes of environmental Vibrio cholerae non-O1, non-O139 strains.
Bhattacharya T, Chatterjee S, Maiti D, Bhadra RK, Takeda Y, Nair GB, Nandy RK. Bhattacharya T, et al. Environ Microbiol. 2006 Mar;8(3):526-634. doi: 10.1111/j.1462-2920.2005.00932.x. Environ Microbiol. 2006. PMID: 16478458 - Phage regulatory circuits and virulence gene expression.
Waldor MK, Friedman DI. Waldor MK, et al. Curr Opin Microbiol. 2005 Aug;8(4):459-65. doi: 10.1016/j.mib.2005.06.001. Curr Opin Microbiol. 2005. PMID: 15979389 Review. - Phage-bacterial interactions in the evolution of toxigenic Vibrio cholerae.
Faruque SM, Mekalanos JJ. Faruque SM, et al. Virulence. 2012 Nov 15;3(7):556-65. doi: 10.4161/viru.22351. Epub 2012 Oct 17. Virulence. 2012. PMID: 23076327 Free PMC article. Review.
Cited by
- Diversity and Complexity of CTXΦ and Pre-CTXΦ Families in Vibrio cholerae from Seventh Pandemic.
Li X, Han Y, Zhao W, Yue X, Huang S, Li Z, Fan F, Liang W, Kan B. Li X, et al. Microorganisms. 2024 Sep 24;12(10):1935. doi: 10.3390/microorganisms12101935. Microorganisms. 2024. PMID: 39458246 Free PMC article. - Recent Vibrio cholerae O1 Epidemic Strains Are Unable To Replicate CTXΦ Prophage Genome.
Ochi K, Mizuno T, Samanta P, Mukhopadhyay AK, Miyoshi SI, Imamura D. Ochi K, et al. mSphere. 2021 Jun 30;6(3):e0033721. doi: 10.1128/mSphere.00337-21. Epub 2021 Jun 9. mSphere. 2021. PMID: 34106768 Free PMC article. - DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae.
Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. Heidelberg JF, et al. Nature. 2000 Aug 3;406(6795):477-83. doi: 10.1038/35020000. Nature. 2000. PMID: 10952301 Free PMC article. - Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease.
Dziejman M, Balon E, Boyd D, Fraser CM, Heidelberg JF, Mekalanos JJ. Dziejman M, et al. Proc Natl Acad Sci U S A. 2002 Feb 5;99(3):1556-61. doi: 10.1073/pnas.042667999. Epub 2002 Jan 29. Proc Natl Acad Sci U S A. 2002. PMID: 11818571 Free PMC article. - VGJ phi, a novel filamentous phage of Vibrio cholerae, integrates into the same chromosomal site as CTX phi.
Campos J, Martínez E, Suzarte E, Rodríguez BL, Marrero K, Silva Y, Ledón T, del Sol R, Fando R. Campos J, et al. J Bacteriol. 2003 Oct;185(19):5685-96. doi: 10.1128/JB.185.19.5685-5696.2003. J Bacteriol. 2003. PMID: 13129939 Free PMC article.
References
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidmann J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1990.
- Basu A, Mukhopadhyay A K, Sharma C, Jyot J, Gupta N, Ghosh A, Bhattacharya S K, Takeda Y, Faruque A S, Albert M J, Balakrish Nair G. Heterogeneity in the organization of the CTX genetic element in strains of Vibrio cholerae O139 Bengal isolated from Calcutta, India and Dhaka, Bangladesh and its possible link to the dissimilar incidence of O139 cholera in the two locales. Microb Pathog. 1998;24:175–183. - PubMed
- Berg, D. Personal communication.
- Boyd, E. F. Unpublished data.
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI042347/AI/NIAID NIH HHS/United States
- P30 DK034928/DK/NIDDK NIH HHS/United States
- AI-42347/AI/NIAID NIH HHS/United States
- P30DK-34928/DK/NIDDK NIH HHS/United States
- T32 AI07329/AI/NIAID NIH HHS/United States
- T32 AI007329/AI/NIAID NIH HHS/United States
- R37 AI042347/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources