Comparative phylogenetic assignment of environmental sequences of genes encoding 16S rRNA and numerically abundant culturable bacteria from an anoxic rice paddy soil - PubMed (original) (raw)
Comparative Study
Comparative phylogenetic assignment of environmental sequences of genes encoding 16S rRNA and numerically abundant culturable bacteria from an anoxic rice paddy soil
U Hengstmann et al. Appl Environ Microbiol. 1999 Nov.
Abstract
We used both cultivation and direct recovery of bacterial 16S rRNA gene (rDNA) sequences to investigate the structure of the bacterial community in anoxic rice paddy soil. Isolation and phenotypic characterization of 19 saccharolytic and cellulolytic strains are described in the accompanying paper (K.-J. Chin, D. Hahn, U. Hengstmann, W. Liesack, and P. H. Janssen, Appl. Environ. Microbiol. 65:5042-5049, 1999). Here we describe the phylogenetic positions of these strains in relation to 57 environmental 16S rDNA clone sequences. Close matches between the two data sets were obtained for isolates from the culturable populations determined by the most-probable-number counting method to be large (3 x 10(7) to 2.5 x 10(8) cells per g [dry weight] of soil). This included matches with 16S rDNA similarity values greater than 98% within distinct lineages of the division Verrucomicrobia (strain PB90-1) and the Cytophaga-Flavobacterium-Bacteroides group (strains XB45 and PB90-2), as well as matches with similarity values greater than 95% within distinct lines of descent of clostridial cluster XIVa (strain XB90) and the family Bacillaceae (strain SB45). In addition, close matches with similarity values greater than 95% were obtained for cloned 16S rDNA sequences and bacteria (strains DR1/8 and RPec1) isolated from the same type of rice paddy soil during previous investigations. The correspondence between culture methods and direct recovery of environmental 16S rDNA suggests that the isolates obtained are representative geno- and phenotypes of predominant bacterial groups which account for 5 to 52% of the total cells in the anoxic rice paddy soil. Furthermore, our findings clearly indicate that a dual approach results in a more objective view of the structural and functional composition of a soil bacterial community than either cultivation or direct recovery of 16S rDNA sequences alone.
Figures
FIG. 1
16S rDNA-based dendrogram showing the phylogenetic relationship of environmental clones BSV64 and BSV69 to MPN isolates PB90-1, PB90-3, and ACB90 (8) and to representative members of the division Verrucomicrobia (21), the order Planctomycetales, and Chlamydia psittaci. Strains VeCb1, VeGlc2, and VeSm13 (underlined) were isolated from Vercelli rice paddy soil in a previous study (25). Based on a 50% invariance criterion for 51 sequences from members of the Verrucomicrobia, Planctomycetales, and the genus Chlamydia, 1,192 nucleotide sequence positions (E. coli 16S rRNA positions 31 to 1476) were used for tree construction. The percentages are the significance values for interior nodes, as derived from a bootstrap test in which 1,000 data resamplings were used. The scale bar represents a 10% estimated difference in nucleotide sequences.
FIG. 2
16S rDNA-based dendrogram showing the phylogenetic relationship of environmental clones (BSV clones) to MPN isolates PB90-2 and XB45 (8) and to representative members of the CFB group. Based on a 50% invariance criterion for 176 sequences from members of the CFB group, 1,190 nucleotide sequence positions (E. coli 16S rRNA positions 110 to 1431) were considered for tree construction. The percentages are the significance values for the interior nodes, as derived from a bootstrap test in which 1,000 data resamplings were used. The root was determined by using the 16S rDNA sequence of E. coli as the outgroup reference sequence. The scale bar represents a 10% estimated difference in nucleotide sequences.
FIG. 3
16S rDNA-based dendrogram showing the phylogenetic relationship of environmental clones (BSV clones) to MPN isolate XB90 (8) and to representative members of clostridial cluster XIVa of Collins et al. (10). Based on a 50% invariance criterion for 326 _Clostridium_-like sequences, 1,238 nucleotide sequence positions (E. coli 16S rRNA positions 31 to 1386) were considered for tree construction. The percentage is the significance value for an interior node, as derived from a bootstrap test in which 1,000 data resamplings were used. The root was determined by using the 16S rDNA sequence of Clostridium fallax as the outgroup reference sequence. The scale bar represents a 10% estimated difference in nucleotide sequences.
FIG. 4
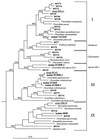
16S rDNA-based dendrogram showing the phylogenetic relationship of environmental clones (BSV clones) to MPN isolates FCB45, FCB90-1, FCB90-2, FCB90-3, SB90, and VeCb10 (8) and to representatives of clostridial clusters I, III, and IX of Collins et al. (10), as well as related groups. Strains RPec1, RCel1, and DR1/8 (underlined) were isolated from Vercelli rice paddy soil in previous studies (9, 38). Based on a 50% invariance criterion for 357 _Clostridium_-like sequences, 1,040 nucleotide sequence positions (E. coli 16S rRNA positions 100 to 1249) were considered for tree construction. The percentages are the significance values for interior nodes, as derived from a bootstrap test in which 1,000 data resamplings were used. The root was determined by using the 16S rDNA sequence of Fervidobacterium islandicum as the outgroup reference sequence. The scale bar represents a 10% estimated difference in nucleotide sequences.
FIG. 5
16S rDNA-based dendrogram showing the phylogenetic relationship of environmental clones (BSV clones) to MPN isolate SB45 (8) and to representative members of the genera Bacillus and Paenibacillus. Based on a 50% invariance criterion for 462 sequences from members of the Bacillaceae, 1,146 nucleotide sequence positions (E. coli 16S rRNA positions 101 to 1274) were considered for tree construction. The percentages are the significance values for interior nodes, as derived from a bootstrap test in which 1,000 data resamplings were used. The root was determined by using the 16S rDNA sequence of Clostridium fallax as the outgroup reference sequence. The scale bar represents a 10% estimated difference in nucleotide sequences.
FIG. 6
16S rDNA-based dendrogram showing the phylogenetic relationship of environmental clone BSV58 to MPN isolates KCB45, PB90-4, PB90-5, VeCb6, and VeSm15 (8) and to representative members of the class Actinobacteria (44). Based on a 50% invariance criterion for 859 sequences from members of the class Actinobacteria, 1,296 nucleotide sequence positions (E. coli 16S rRNA positions 41 to 1375) were considered for tree construction (except for isolate KCB45). Isolate KCB45 was inserted into the tree based on a continuous sequence stretch corresponding to E. coli 16S rRNA positions 41 to 1060. The percentages are the significance values for interior nodes, as derived from a bootstrap test in which 1,000 data resamplings were used. The root was determined by using the 16S rDNA sequence of Clostridium fallax as the outgroup reference sequence. The scale bar represents a 10% estimated difference in nucleotide sequences.
Similar articles
- Novel bacterial lineages at the (sub)division level as detected by signature nucleotide-targeted recovery of 16S rRNA genes from bulk soil and rice roots of flooded rice microcosms.
Derakshani M, Lukow T, Liesack W. Derakshani M, et al. Appl Environ Microbiol. 2001 Feb;67(2):623-31. doi: 10.1128/AEM.67.2.623-631.2001. Appl Environ Microbiol. 2001. PMID: 11157225 Free PMC article. - Characterization and identification of numerically abundant culturable bacteria from the anoxic bulk soil of rice paddy microcosms.
Chin KJ, Hahn D, Hengstmann U, Liesack W, Janssen PH. Chin KJ, et al. Appl Environ Microbiol. 1999 Nov;65(11):5042-9. doi: 10.1128/AEM.65.11.5042-5049.1999. Appl Environ Microbiol. 1999. PMID: 10543821 Free PMC article. - Bacterial populations colonizing and degrading rice straw in anoxic paddy soil.
Weber S, Stubner S, Conrad R. Weber S, et al. Appl Environ Microbiol. 2001 Mar;67(3):1318-27. doi: 10.1128/AEM.67.3.1318-1327.2001. Appl Environ Microbiol. 2001. PMID: 11229927 Free PMC article. - Hymenobacter roseosalivarius gen. nov., sp. nov. from continental Antartica soils and sandstone: bacteria of the Cytophaga/Flavobacterium/Bacteroides line of phylogenetic descent.
Hirsch P, Ludwig W, Hethke C, Sittig M, Hoffmann B, Gallikowski CA. Hirsch P, et al. Syst Appl Microbiol. 1998 Aug;21(3):374-383. doi: 10.1016/s0723-2020(98)80047-7. Syst Appl Microbiol. 1998. PMID: 9841127 - How many species of prokaryotes are there?
Ward BB. Ward BB. Proc Natl Acad Sci U S A. 2002 Aug 6;99(16):10234-6. doi: 10.1073/pnas.162359199. Epub 2002 Jul 30. Proc Natl Acad Sci U S A. 2002. PMID: 12149517 Free PMC article. Review. No abstract available.
Cited by
- Phylogenetic analysis of anaerobic psychrophilic enrichment cultures obtained from a greenland glacier ice core.
Sheridan PP, Miteva VI, Brenchley JE. Sheridan PP, et al. Appl Environ Microbiol. 2003 Apr;69(4):2153-60. doi: 10.1128/AEM.69.4.2153-2160.2003. Appl Environ Microbiol. 2003. PMID: 12676695 Free PMC article. - Populations implicated in anaerobic reductive dechlorination of 1,2-dichloropropane in highly enriched bacterial communities.
Ritalahti KM, Löffler FE. Ritalahti KM, et al. Appl Environ Microbiol. 2004 Jul;70(7):4088-95. doi: 10.1128/AEM.70.7.4088-4095.2004. Appl Environ Microbiol. 2004. PMID: 15240287 Free PMC article. - Phylogenetic evidence of noteworthy microflora from the subsurface of the former Homestake gold mine, Lead, South Dakota.
Waddell EJ, Elliott TJ, Vahrenkamp JM, Roggenthen WM, Sani RK, Anderson CM, Bang SS. Waddell EJ, et al. Environ Technol. 2010 Jul-Aug;31(8-9):979-91. doi: 10.1080/09593331003789511. Environ Technol. 2010. PMID: 20662386 Free PMC article. - Isolation and biodiversity of hitherto undescribed soil bacteria related to Bacillus niacini.
Felske AD, Tzeneva V, Heyrman J, Langeveld MA, Akkermans AD, De Vos P. Felske AD, et al. Microb Ecol. 2004 Jul;48(1):111-9. doi: 10.1007/s00248-003-2025-4. Epub 2004 Mar 25. Microb Ecol. 2004. PMID: 15037965 - Novel bacterial lineages at the (sub)division level as detected by signature nucleotide-targeted recovery of 16S rRNA genes from bulk soil and rice roots of flooded rice microcosms.
Derakshani M, Lukow T, Liesack W. Derakshani M, et al. Appl Environ Microbiol. 2001 Feb;67(2):623-31. doi: 10.1128/AEM.67.2.623-631.2001. Appl Environ Microbiol. 2001. PMID: 11157225 Free PMC article.
References
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1995.
- Bakken L R, Olsen R A. The relationship between cell size and viability of soil bacteria. Microb Ecol. 1987;13:103–114. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases