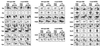Role of CcpA in regulation of the central pathways of carbon catabolism in Bacillus subtilis - PubMed (original) (raw)
Role of CcpA in regulation of the central pathways of carbon catabolism in Bacillus subtilis
S Tobisch et al. J Bacteriol. 1999 Nov.
Abstract
The Bacillus subtilis two-dimensional (2D) protein index contains almost all glycolytic and tricarboxylic acid (TCA) cycle enzymes, among them the most abundant housekeeping proteins of growing cells. Therefore, a comprehensive study on the regulation of glycolysis and the TCA cycle was initiated. Whereas expression of genes encoding the upper and lower parts of glycolysis (pgi, pfk, fbaA, and pykA) is not affected by the glucose supply, there is an activation of the glycolytic gap gene and the pgk operon by glucose. This activation seems to be dependent on the global regulator CcpA, as shown by 2D polyacrylamide gel electrophoresis analysis as well as by transcriptional analysis. Furthermore, a high glucose concentration stimulates production and excretion of organic acids (overflow metabolism) in the wild type but not in the ccpA mutant. Finally, CcpA is involved in strong glucose repression of almost all TCA cycle genes. In addition to TCA cycle and glycolytic enzymes, the levels of many other proteins are affected by the ccpA mutation. Our data suggest (i) that ccpA mutants are unable to activate glycolysis or carbon overflow metabolism and (ii) that CcpA might be a key regulator molecule, controlling a superregulon of glucose catabolism.
Figures
FIG. 1
Main pathways of carbohydrate metabolism. (A) Silver-stained 2D gel showing identified proteins which are involved in glycolysis (Pgi, Pfk, FbaA, Tpi, Gap, Pgk, Pgm, Eno), pyruvate dehydrogenesis (PdhA, PdhB, PdhC, PdhD), the TCA cycle (CitZ, CitB, CitC, OdhA, PdhD, SucC, SucD, SdhA, CitG, CitH), and overflow metabolism (Pta, AckA). (B) Schematic representation of glycolysis and the TCA cycle. Enzymes and metabolites are indicated. Note that not all the proteins of these pathways are identified on 2D gels.
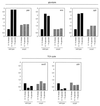
FIG. 2
Comparison of CcpA-dependent expression of proteins involved in glycolysis (□), the TCA cycle (○), and overflow metabolism (◊) in B. subtilis. The spots correspond to enzymes of central carbon metabolism. They are derived from silver-stained 2D electrophoretograms of B. subtilis IS58 and the ccpA mutant strain BGW2, which were grown in pure ASM without any additional carbon source or with 1% glucose.
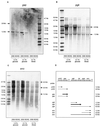
FIG. 3
Northern blot analysis of genes encoding glycolytic enzymes. RNA of exponentially growing B. subtilis cells was hybridized with probes specific for gap (A), pgk (B), and eno (C). Sizes of transcripts are indicated, and possible interpretations are illustrated (D); thicknesses of arrows correspond with the amount of transcript.
FIG. 4
Quantitative analysis of mRNAs of glycolytic and TCA cycle enzymes. Different concentrations of RNA (1 and 0.5 μg) from B. subtilis IS58 and BGW2 were blotted onto nylon membranes and hybridized with probes specific for the genes indicated. The mRNA amount obtained for IS58 in the presence of 0.1% ribose was set at 1.
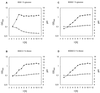
FIG. 5
CcpA- and carbon source-dependent acidification of growth medium. Measurement of pH (□) during growth (⧫) was carried out as described in Materials and Methods for the B. subtilis IS58 wild-type strain in ASM containing 1% glucose (A) or 0.1% ribose (B), as well as for the B. subtilis BGW2 ccpA mutant strain in ASM containing 1% glucose (C) or 0.1% ribose (D).
FIG. 6
Silver-stained 2D gels of B. subtilis wild-type strain IS58 (A) and isogenic ccpA mutant strain BGW2 (B) protein extracts. Glycolytic enzymes (□), enzymes of the TCA cycle (○), and enzymes of overflow metabolism (◊) are indicated, as are proteins which are repressed ( ) or induced (
) or induced ( ) in the absence of CcpA.
) in the absence of CcpA.
Similar articles
- Regulation of pho regulon gene expression by the carbon control protein A, CcpA, in Bacillus subtilis.
Choi SK, Saier MH Jr. Choi SK, et al. J Mol Microbiol Biotechnol. 2005;10(1):40-50. doi: 10.1159/000090347. J Mol Microbiol Biotechnol. 2005. PMID: 16491025 - Carbon catabolite control of the metabolic network in Bacillus subtilis.
Fujita Y. Fujita Y. Biosci Biotechnol Biochem. 2009 Feb;73(2):245-59. doi: 10.1271/bbb.80479. Epub 2009 Feb 7. Biosci Biotechnol Biochem. 2009. PMID: 19202299 Review. - Bacillus subtilis ccpA gene mutants specifically defective in activation of acetoin biosynthesis.
Turinsky AJ, Moir-Blais TR, Grundy FJ, Henkin TM. Turinsky AJ, et al. J Bacteriol. 2000 Oct;182(19):5611-4. doi: 10.1128/JB.182.19.5611-5614.2000. J Bacteriol. 2000. PMID: 10986270 Free PMC article. - Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA.
Luesink EJ, van Herpen RE, Grossiord BP, Kuipers OP, de Vos WM. Luesink EJ, et al. Mol Microbiol. 1998 Nov;30(4):789-98. doi: 10.1046/j.1365-2958.1998.01111.x. Mol Microbiol. 1998. PMID: 10094627 - The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis.
Henkin TM. Henkin TM. FEMS Microbiol Lett. 1996 Jan 1;135(1):9-15. doi: 10.1111/j.1574-6968.1996.tb07959.x. FEMS Microbiol Lett. 1996. PMID: 8598282 Review.
Cited by
- Influence of the Alternative Sigma Factor RpoN on Global Gene Expression and Carbon Catabolism in Enterococcus faecalis V583.
Keffeler EC, Iyer VS, Parthasarathy S, Ramsey MM, Gorman MJ, Barke TL, Varahan S, Olson S, Gilmore MS, Abdullahi ZH, Hancock EN, Hancock LE. Keffeler EC, et al. mBio. 2021 May 18;12(3):e00380-21. doi: 10.1128/mBio.00380-21. mBio. 2021. PMID: 34006651 Free PMC article. - Bacillus subtilis metabolism and energetics in carbon-limited and excess-carbon chemostat culture.
Dauner M, Storni T, Sauer U. Dauner M, et al. J Bacteriol. 2001 Dec;183(24):7308-17. doi: 10.1128/JB.183.24.7308-7317.2001. J Bacteriol. 2001. PMID: 11717290 Free PMC article. - Overexpression of PrfA leads to growth inhibition of Listeria monocytogenes in glucose-containing culture media by interfering with glucose uptake.
Marr AK, Joseph B, Mertins S, Ecke R, Müller-Altrock S, Goebel W. Marr AK, et al. J Bacteriol. 2006 Jun;188(11):3887-901. doi: 10.1128/JB.01978-05. J Bacteriol. 2006. PMID: 16707681 Free PMC article. - Stochasticity versus determinism: consequences for realistic gene regulatory network modelling and evolution.
Jenkins DJ, Stekel DJ. Jenkins DJ, et al. J Mol Evol. 2010 Feb;70(2):215-31. doi: 10.1007/s00239-010-9323-5. Epub 2010 Feb 12. J Mol Evol. 2010. PMID: 20151115 - Comparison of the regulation, metabolic functions, and roles in virulence of the glyceraldehyde-3-phosphate dehydrogenase homologues gapA and gapB in Staphylococcus aureus.
Purves J, Cockayne A, Moody PC, Morrissey JA. Purves J, et al. Infect Immun. 2010 Dec;78(12):5223-32. doi: 10.1128/IAI.00762-10. Epub 2010 Sep 27. Infect Immun. 2010. PMID: 20876289 Free PMC article.
References
- Antelmann H, Bernhardt J, Schmid R, Mach H, Völker U, Hecker M. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis. 1997;18:1451–1463. - PubMed
- Aymerich, S. Personal communication.
- Bernhardt J, Völker U, Völker A, Antelmann H, Schmid R, Mach H, Hecker M. Specific and general stress proteins in Bacillus subtilis—a two dimensional protein electrophoresis study. Microbiology. 1997;143:999–1017. - PubMed
- Berhardt J, Werner H. revision date. The 2D protein index of Bacillus subtilis—Sub2D. 19 August 1999. http://microbiol.biologie.uni-greifswald.de::88801 [Online.] http://microbiol.biologie.uni-greifswald.de::88801. Ernst-Moritz-Arndt-Universität Greifswald, Griefswald, Germany. . Ernst-Moritz-Arndt-Universitt Greifswald, Griefswald, Germany.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous