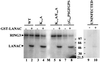Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene - PubMed (original) (raw)
Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene
G M Platt et al. J Virol. 1999 Dec.
Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) is the likely infectious cause of Kaposi's sarcoma, primary effusion lymphoma, and some cases of multicentric Castleman's disease. Its latent nuclear antigen (LANA) is expressed in the nuclei of latently infected cells and may play a role in the persistence of episomal viral DNA in dividing cells. Here we report that LANA interacts with RING3, a nuclear protein and member of the Drosophila fsh (female sterile homeotic) family of proteins, some of which have previously been implicated in controlling gene expression. Binding of RING3 to LANA involves the ET domain, characteristic of fsh-related proteins, suggesting that this highly conserved region is involved in protein-protein interactions. The interaction between RING3 and LANA results in phosphorylation of serine and threonine residues located between amino acids 951 and 1107 in the carboxy-terminal region of LANA. However, RING3 is not itself a kinase but appears to recruit an as yet unidentified serine/threonine protein kinase into the complex which it forms with LANA.
Figures
FIG. 1
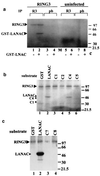
Interaction of RING3 and LANA. (a) RING3 binds to the C-terminal 212 amino acids of LANA. Lysates of uninfected Sf9 cells (lanes 1 and 3) or Sf9 cells infected with the recombinant RING3 baculovirus (lanes 2 and 4) were incubated with either GST-LANAC (lanes 1 and 2) or GST alone (lanes 3 and 4) immobilized on glutathione beads. Binding of RING3 was detected by Western blot analysis with an affinity-purified rabbit antibody raised against the R3B fusion protein (Fig. 3). (b) The complete LANA protein from the BCP-1 cell line binds to RING3. Cellular lysates of BCP-1 cells were incubated with a GST RING3 fusion protein (amino acids 554 to 754) (lane 1) or GST alone (lane 2). Binding of LANA was detected on Western blots with serum from a KS patient. (c) Detection of LANA-RING3 complexes in BCP-1 cells by coimmunoprecipitation. Lysates of BCP-1 cells were incubated with either the RING3 antibody (lanes 2 and 3) or prebleed (pb) rabbit serum (lanes 1 and 4). Immunoprecipitation (IP) with a rabbit antibody raised against the C-terminal LANA region was used as a positive control for the presence of LANA (lane 5). The protein-antibody complexes were isolated by using protein G beads and analyzed by Western blotting with either a rat anti-RING3 peptide antibody (a gift from K. Thorpe, Cambridge University, Cambridge, United Kingdom) (lanes 1 and 2) or human serum from a KS patient (lanes 3 to 5). The positions of the RING3 and LANA proteins are indicated. The sizes of the protein markers are shown in kilodaltons.
FIG. 2
Deletion mapping of the LANA domains which interact with RING3. (a) Schematic diagram of the LANA fragments expressed as fusion proteins. The ability to bind to RING3 is summarized on the right. The positions of the internal repeat region (repeat), the nuclear localization signal (NLS), and the leucine zipper motif within LANA are indicated. LANA is numbered 1 to 1162 as in a previous study (25, 27). (b and c) Lysates prepared from Sf9 cells infected with the RING3 recombinant baculovirus were incubated with different GST-LANA deletion proteins, and binding was detected by using the affinity-purified antibody to RING3. (d and e) The GST-LANA fusion proteins used in the binding on a Coomassie blue-stained SDS-polyacrylamide gel.
FIG. 3
Deletion mapping of RING3 domains which interact with LANA. (a) Schematic diagram of the RING3 regions used in the construction of the GST-RING3 deletion proteins. The RING3 protein is numbered 1 to 754 as in a previous study (3). The positions of the two extended bromodomains (15), the ET domain (21), the nuclear localization signal (NLS), the postulated ATP binding motif (G554PSGFGPSG561), the catalytic lysine (K574), and the catalytic glutamic acid (E599) (8) are indicated. The ability of the GST RING3 fusion proteins to bind to LANA is summarized on the right. (b and c) Binding of LANA to the RING3 deletion proteins. Lysates of Sf9 cells expressing the recombinant c-terminal LANA fragment (LANAC) (b) or complete LANA (c) were incubated with the GST-RING3 deletion proteins immobilized on glutathione-Sepharose beads, and binding was detected on Western blots with serum from a patient with KS. The sizes of the protein markers are indicated in kilodaltons on the left. (d) The GST-RING3 fusion proteins used in the binding assays on a Coomassie blue-stained SDS-polyacrylamide gel.
FIG. 4
The C-terminal domain of LANA becomes phosphorylated in the presence of RING3. (a) Cellular lysates of SF9 cells infected with the RING3 baculovirus (lanes 1 to 4) or uninfected SF9 cells (lanes 5 to 8) were immunoprecipitated (IP) with either a rabbit antibody to RING3 (lanes 1, 2, 5, and 6) or prebleed (pb) rabbit serum (lanes 3, 4, 7, and 8). The immunoprecipitated proteins were added to an in vitro kinase assay mixture containing either 500 ng of GST-LANAC (lanes 2, 4, 6, and 8) or 500 ng of GST alone (lanes 1, 3, 5, and 7) as substrates and [γ-32P]ATP. Phosphorylated proteins were resolved by SDS-polyacrylamide gel electrophoresis and visualized by autoradiography. The positions of the phosphorylated RING3 and GST-LANAC proteins are indicated, and the sizes of the protein markers are indicated in kilodaltons on the right. (b) A 500-ng portion of each of the GST LANA deletion proteins C1 (lane 3), C2 (lane 4), C4 (lane 5), and C5 (lane 6) (Fig. 2) was used as a substrate in an in vitro kinase assay in the presence of the recombinant RING3 protein. GST (lane 1) or GST-LANAC (lane 2) were used as negative and positive controls, respectively. (c) A 500-ng portion of each of the GST-LANA deletion proteins C7 (lane 3) and C8 (lane 4) were used as substrates in an in vitro kinase assay.
FIG. 5
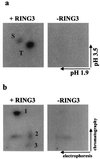
Phosphorylation of LANA in the presence of RING3 occurs on specific sites. GST-LANAC was phosphorylated by an in vitro kinase assay in the presence or absence of immunoprecipitated recombinant RING3, as in Fig. 4, and eluted from a gel slice (38). (a) Phosphoamino acid analysis by two-dimensional electrophoresis on cellulose-acetate sheets. The positions of the phosphoserine (S) and phosphothreonine (T) and the direction of the first (pH 1.9) and second (pH 3.5) electrophoretic dimensions are indicated. (b) Tryptic phosphopeptide map. Tryptic peptides were separated on thin-layer chromatography plates. The directions of the first (electrophoresis) and second (chromatography) dimensions are indicated. The numbers 1 to 3 denote the specific peptides observed only in the presence of immunoprecipitated RING3.
FIG. 6
Mutational analysis of the putative protein kinase subdomains of RING3. Three mutant, insect cell-expressed, immunoprecipitated RING3 proteins were added to an in vitro kinase assay mixture in the presence of either GST (lanes 1, 3, 5, and 7) or GST-LANAC (lanes 2, 4, 6, and 8), as in Fig. 4. Wild-type RING3 protein (WT) (lanes 1 and 2), mutation of the postulated catalytic lysine residue to alanine (K574A) (lanes 3 and 4), mutation of the two postulated catalytic residues K574 and E599 to alanine (K574/E599) (lanes 5 and 6), deletion of 8 amino acids within the postulated ATP binding motif (ΔG554PSGFGPS561) (lanes 7 and 8), and uninfected Sf9 lysates (lanes 9 and 10) are shown.
Similar articles
- Latent nuclear antigen of Kaposi's sarcoma herpesvirus/human herpesvirus-8 induces and relocates RING3 to nuclear heterochromatin regions.
Mattsson K, Kiss C, Platt GM, Simpson GR, Kashuba E, Klein G, Schulz TF, Szekely L. Mattsson K, et al. J Gen Virol. 2002 Jan;83(Pt 1):179-188. doi: 10.1099/0022-1317-83-1-179. J Gen Virol. 2002. PMID: 11752715 - Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's Sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1.
Viejo-Borbolla A, Ottinger M, Brüning E, Bürger A, König R, Kati E, Sheldon JA, Schulz TF. Viejo-Borbolla A, et al. J Virol. 2005 Nov;79(21):13618-29. doi: 10.1128/JVI.79.21.13618-13629.2005. J Virol. 2005. PMID: 16227282 Free PMC article. - Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest.
Ottinger M, Christalla T, Nathan K, Brinkmann MM, Viejo-Borbolla A, Schulz TF. Ottinger M, et al. J Virol. 2006 Nov;80(21):10772-86. doi: 10.1128/JVI.00804-06. Epub 2006 Aug 23. J Virol. 2006. PMID: 16928766 Free PMC article. - The Kaposi' s sarcoma-associated herpesvirus latency-associated nuclear antigen.
Komatsu T, Barbera AJ, Ballestas ME, Kaye KM. Komatsu T, et al. Viral Immunol. 2001;14(4):311-7. doi: 10.1089/08828240152716565. Viral Immunol. 2001. PMID: 11792061 Review. - Pathological Features of Kaposi's Sarcoma-Associated Herpesvirus Infection.
Katano H. Katano H. Adv Exp Med Biol. 2018;1045:357-376. doi: 10.1007/978-981-10-7230-7_16. Adv Exp Med Biol. 2018. PMID: 29896675 Review.
Cited by
- Enhancer-promoter activation by the Kaposi sarcoma-associated herpesvirus episome maintenance protein LANA.
Ye X, Guerin LN, Chen Z, Rajendren S, Dunker W, Zhao Y, Zhang R, Hodges E, Karijolich J. Ye X, et al. Cell Rep. 2024 Mar 26;43(3):113888. doi: 10.1016/j.celrep.2024.113888. Epub 2024 Feb 27. Cell Rep. 2024. PMID: 38416644 Free PMC article. - Bromodomain and BET family proteins as epigenetic targets in cancer therapy: their degradation, present drugs, and possible PROTACs.
Muddassir M, Soni K, Sangani CB, Alarifi A, Afzal M, Abduh NAY, Duan Y, Bhadja P. Muddassir M, et al. RSC Adv. 2020 Dec 24;11(2):612-636. doi: 10.1039/d0ra07971e. eCollection 2020 Dec 24. RSC Adv. 2020. PMID: 35746919 Free PMC article. Review. - Relevance of BET Family Proteins in SARS-CoV-2 Infection.
Lara-Ureña N, García-Domínguez M. Lara-Ureña N, et al. Biomolecules. 2021 Jul 30;11(8):1126. doi: 10.3390/biom11081126. Biomolecules. 2021. PMID: 34439792 Free PMC article. Review. - Brd/BET Proteins Influence the Genome-Wide Localization of the Kaposi's Sarcoma-Associated Herpesvirus and Murine Gammaherpesvirus Major Latency Proteins.
Lotke R, Schneeweiß U, Pietrek M, Günther T, Grundhoff A, Weidner-Glunde M, Schulz TF. Lotke R, et al. Front Microbiol. 2020 Oct 22;11:591778. doi: 10.3389/fmicb.2020.591778. eCollection 2020. Front Microbiol. 2020. PMID: 33193257 Free PMC article. - LANA oligomeric architecture is essential for KSHV nuclear body formation and viral genome maintenance during latency.
De Leo A, Deng Z, Vladimirova O, Chen HS, Dheekollu J, Calderon A, Myers KA, Hayden J, Keeney F, Kaufer BB, Yuan Y, Robertson E, Lieberman PM. De Leo A, et al. PLoS Pathog. 2019 Jan 25;15(1):e1007489. doi: 10.1371/journal.ppat.1007489. eCollection 2019 Jan. PLoS Pathog. 2019. PMID: 30682185 Free PMC article.
References
- Ballestas M E, Chatis P A, Kaye K H. Efficient persistance of extrachromosomal KSHV DNA mediated by latency associated nuclear antigen. Science. 1999;284:641–644. - PubMed
- Beck S, Hanson I, Kelly A, Pappin D J, Trowsdale J. A homologue of the Drosophila female sterile homeotic (fsh) gene in the class II region of the human MHC. DNA Seq. 1992;2:203–210. - PubMed
- Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O’Leary J J. Kaposi’s sarcoma associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1:1274–1278. - PubMed
- Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous