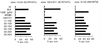Efficient processing of the immunodominant, HLA-A*0201-restricted human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitope despite multiple variations in the epitope flanking sequences - PubMed (original) (raw)
. 1999 Dec;73(12):10191-8.
doi: 10.1128/JVI.73.12.10191-10198.1999.
O O Yang, N G Jones, Y Lee, P Goulder, R P Johnson, A Trocha, D Colbert, C Hay, S Buchbinder, C C Bergmann, H J Zweerink, S Wolinsky, W A Blattner, S A Kalams, B D Walker
Affiliations
- PMID: 10559335
- PMCID: PMC113072
- DOI: 10.1128/JVI.73.12.10191-10198.1999
Efficient processing of the immunodominant, HLA-A*0201-restricted human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitope despite multiple variations in the epitope flanking sequences
C Brander et al. J Virol. 1999 Dec.
Abstract
Immune escape from cytotoxic T-lymphocyte (CTL) responses has been shown to occur not only by changes within the targeted epitope but also by changes in the flanking sequences which interfere with the processing of the immunogenic peptide. However, the frequency of such an escape mechanism has not been determined. To investigate whether naturally occurring variations in the flanking sequences of an immunodominant human immunodeficiency virus type 1 (HIV-1) Gag CTL epitope prevent antigen processing, cells infected with HIV-1 or vaccinia virus constructs encoding different patient-derived Gag sequences were tested for recognition by HLA-A*0201-restricted, p17-specific CTL. We found that the immunodominant p17 epitope (SL9) and its variants were efficiently processed from minigene expressing vectors and from six HIV-1 Gag variants expressed by recombinant vaccinia virus constructs. Furthermore, SL9-specific CTL clones derived from multiple donors efficiently inhibited virus replication when added to HLA-A*0201-bearing cells infected with primary or laboratory-adapted strains of virus, despite the variability in the SL9 flanking sequences. These data suggest that escape from this immunodominant CTL response is not frequently accomplished by changes in the epitope flanking sequences.
Figures
FIG. 1
Recognition of the SL9 epitope expressed by different vaccinia virus constructs. HLA-A*0201-positive B-LCL cells were infected overnight with vaccinia virus constructs or pulsed with optimal peptides (SL9 for clones 115.D4 and 13010.B17) or, in this experiment, with the HLA-B52 restricted peptide 122E (Gag p24, RMYSPTSI, amino acids 275 to 282) recognized by clone 19-203 for 90 min. The effector/target ratio was 5:1. This experiment was repeated three times with different CTL clones and target cell lines from three different donors, yielding the same pattern of recognition.
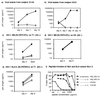
FIG. 2
Processing of SL9 from patient LWF-derived Gag sequences expressed as vaccinia virus minigene constructs and episomal vectors. (A) Vaccinia virus construct expression system. HLA-A*0201-positive B-LCL cells were infected overnight with vaccinia virus constructs expressing patient LWF-derived p17 sequences or the SL9 epitope only. (B) Episomal vector expression system. Alternatively, HMY-A2 cells were stably transfected with episomal vectors expressing SL9 with or without three additional N-terminal amino acid residues. B-LCL and HMY-A2 target cells were 51Cr labeled for 90 min and incubated with a polyclonal SL9-specific CTL line at the indicated effector/target ratios for 4 h in a standard cytotoxicity assay.
FIG. 3
Inhibition of viral replication by SL9-specific CTL clones and peptide titration of SL9 and the NL4-3 encoded variant V821/T84V. (a and b) HLA-A*0201-expressing CD4 cells were infected with viral isolates from subjects VI-06 or 053i and cultured in the presence (◊) or absence (⧫) of the SL9-specific CTL cone 115D4. (c and d) HLA-A*0201-positive T1 cells (c) and HLA-A*0201-negative H9 cells (d) were infected with HIV-1 IIIB and cultured in the presence of SL9-specific CTL clone 115D4 (◊) or clone 161jXA/14 (▵) or without CTL (⧫). (e) T1 cells were infected with HIV-1 NL4-3 isolate and cultured with the same clones as for panels c and d. The production of p24 was measured after 3 to 15 days. (f) For peptide titrations, clones 115.D4 and 161j.XA/14 were both tested on the SL9 sequences expressed by HIV-1 HXB2 (HIV-1 IIIB, SL9 consensus sequence) and by HIV-1 NL4-3 (SL9 variant V82I/T84V). Peptide titration was carried out with HLA-A*0201-positive EBV-transformed B-LCL cells as target cells in a standard 51Cr release assay.
Similar articles
- Impaired processing and presentation of cytotoxic-T-lymphocyte (CTL) epitopes are major escape mechanisms from CTL immune pressure in human immunodeficiency virus type 1 infection.
Yokomaku Y, Miura H, Tomiyama H, Kawana-Tachikawa A, Takiguchi M, Kojima A, Nagai Y, Iwamoto A, Matsuda Z, Ariyoshi K. Yokomaku Y, et al. J Virol. 2004 Feb;78(3):1324-32. doi: 10.1128/jvi.78.3.1324-1332.2004. J Virol. 2004. PMID: 14722287 Free PMC article. - Recognition patterns of HLA-A2-restricted human immunodeficiency virus-1-specific cytotoxic T-lymphocytes in a cohort of HIV-1-infected individuals.
Schmitt-Haendle M, Bachmann O, Harrer E, Schmidt B, Bäuerle M, Harrer T. Schmitt-Haendle M, et al. Viral Immunol. 2005;18(4):627-36. doi: 10.1089/vim.2005.18.627. Viral Immunol. 2005. PMID: 16359229 - The HIV-1 HLA-A2-SLYNTVATL is a help-independent CTL epitope.
Kan-Mitchell J, Bisikirska B, Wong-Staal F, Schaubert KL, Bajcz M, Bereta M. Kan-Mitchell J, et al. J Immunol. 2004 May 1;172(9):5249-61. doi: 10.4049/jimmunol.172.9.5249. J Immunol. 2004. PMID: 15100263 - Escape of human immunodeficiency virus from immune control.
McMichael AJ, Phillips RE. McMichael AJ, et al. Annu Rev Immunol. 1997;15:271-96. doi: 10.1146/annurev.immunol.15.1.271. Annu Rev Immunol. 1997. PMID: 9143689 Review. - The influence of HLA/HIV genetics on the occurrence of elite controllers and a need for therapeutics geotargeting view.
Lunardi LW, Bragatte MAS, Vieira GF. Lunardi LW, et al. Braz J Infect Dis. 2021 Sep-Oct;25(5):101619. doi: 10.1016/j.bjid.2021.101619. Epub 2021 Sep 22. Braz J Infect Dis. 2021. PMID: 34562387 Free PMC article. Review.
Cited by
- Diversity of escape variant mutations in Simian virus 40 large tumor antigen (SV40 Tag) epitopes selected by cytotoxic T lymphocyte (CTL) clones.
Mylin LM, Schell TD, Epler M, Kusuma C, Assis D, Matsko C, Smith A, Allebach A, Tevethia SS. Mylin LM, et al. Virology. 2007 Jul 20;364(1):155-68. doi: 10.1016/j.virol.2007.02.007. Epub 2007 Mar 21. Virology. 2007. PMID: 17368499 Free PMC article. - Intrapatient escape in the A*0201-restricted epitope SLYNTVATL drives evolution of human immunodeficiency virus type 1 at the population level.
Edwards CT, Pfafferott KJ, Goulder PJ, Phillips RE, Holmes EC. Edwards CT, et al. J Virol. 2005 Jul;79(14):9363-6. doi: 10.1128/JVI.79.14.9363-9366.2005. J Virol. 2005. PMID: 15994836 Free PMC article. - The evolutionary significance of certain amino acid substitutions and their consequences for HIV-1 immunogenicity toward HLA's A*0201 and B*27.
Hecht L, Dormer A. Hecht L, et al. Bioinformation. 2013 Mar 19;9(6):315-20. doi: 10.6026/97320630009315. Print 2013. Bioinformation. 2013. PMID: 23745018 Free PMC article. - Formaldehyde-treated, heat-inactivated virions with increased human immunodeficiency virus type 1 env can be used to induce high-titer neutralizing antibody responses.
Poon B, Hsu JF, Gudeman V, Chen IS, Grovit-Ferbas K. Poon B, et al. J Virol. 2005 Aug;79(16):10210-7. doi: 10.1128/JVI.79.16.10210-10217.2005. J Virol. 2005. PMID: 16051814 Free PMC article. - Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection.
Goulder PJ, Altfeld MA, Rosenberg ES, Nguyen T, Tang Y, Eldridge RL, Addo MM, He S, Mukherjee JS, Phillips MN, Bunce M, Kalams SA, Sekaly RP, Walker BD, Brander C. Goulder PJ, et al. J Exp Med. 2001 Jan 15;193(2):181-94. doi: 10.1084/jem.193.2.181. J Exp Med. 2001. PMID: 11148222 Free PMC article.
References
- Androlewicz M J, Cresswell P. How selective is the transporter associated with antigen processing? Immunity. 1996;5:1–5. - PubMed
- Bergmann C C, Yao Q, Ho C, Buckwold S L. Flanking residues alter antigenicity and immunogenicity of multi-unit CTL epitopes. J Immunol. 1996;157:3242–3249. - PubMed
- Borrow P, Lewicki H, Wei X, Horwitz M S, Peffer N, Meyers H, Nelson J A, Gairin J E, Hahn B H, Oldstone M B, Shaw G M. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. - PubMed
- Brander C, Hartman K E, Trocha A K, Jones N G, Johnson R P, Korber B, Wentworth P, Buchbinder S P, Wolinsky S, Walker B D, Kalams S A. Lack of strong immune selection pressure by the immunodominant, HLA-A*0201-restricted CTL response in chronic HIV-1 infection. J Clin Investig. 1998;101:2559–2566. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI33314/AI/NIAID NIH HHS/United States
- R37 AI028568/AI/NIAID NIH HHS/United States
- AI33327/AI/NIAID NIH HHS/United States
- R01 AI039966/AI/NIAID NIH HHS/United States
- R01 AI028568/AI/NIAID NIH HHS/United States
- R01 AI030914/AI/NIAID NIH HHS/United States
- HD31756/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials