Efficient trans-complementation of the flavivirus kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase - PubMed (original) (raw)
Efficient trans-complementation of the flavivirus kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase
A A Khromykh et al. J Virol. 1999 Dec.
Abstract
Successful trans-complementation of the defective Kunjin virus (KUN) RNA FLdGDD with a deletion of the RNA polymerase motif GDD in the NS5 gene by using a BHK cell line, repBHK, that continuously produced a functionally active KUN replication complex (RC) from replicon RNA was recently reported (A. A. Khromykh, M. T. Kenney, and E. G. Westaway, J. Virol. 72:7270-7279, 1998). In order to identify whether this complementation of FLdGDD RNA was provided by the wild-type NS5 protein alone or with the help of other nonstructural (NS) proteins also expressed in repBHK cells, we generated BHK cell lines stably producing the individual NS5 protein (SRns5BHK) or the NS1-NS5 polyprotein (SRns1-5BHK) by using a heterologous expression vector based on a modified noncytopathic Sindbis replicon. Western blot analysis with anti-NS5 antibodies showed that the level of production of NS5 was significantly higher in SRns5BHK cells than in SRns1-5BHK cells. Despite the higher level of expressed NS5, trans-complementation of FLdGDD RNA was much less efficient in SRns5BHK cells than in SRns1-5BHK cells and produced at least 100-fold less of the secreted complemented virus. In contrast, efficient complementation of KUN RNA with lethal cysteine-to-alanine mutations in the NS1 gene was achieved both in BHK cells producing the individual KUN NS1 protein from the Sindbis replicon vector and in repBHK cells, with both cell lines expressing similar amounts of NS1 protein. These results clearly demonstrate that flavivirus NS5 coexpressed with other components of the viral replicase possesses much higher functional (trans-complementing) activity than individually expressed NS5 and that efficient trans-complementation of mutated flavivirus NS1 and NS5 proteins occurs by different mechanisms. The results are interpreted and discussed in relation to our proposed model of formation of the flavivirus RC largely based on previous ultrastructural and biochemical analyses of KUN replication.
Figures
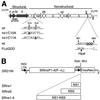
FIG. 1
(A) Schematic representation of mutated KUN cDNA constructs ns1/C10A and ns1/C11A for NS1 and FLd_GDD_ for NS5. The wild type (wt) represents the native KUN sequence flanking the mutated cysteine (
C
) residue numbers 10 and 11 (indicated by arrows) in NS1 and flanking the GDD motif in NS5. Cysteine residues were mutated to alanine (
A
) by PCR mutagenesis of the NS1 gene as described in Materials and Methods. Construction of FLd_GDD_ cDNA was described previously (14). Numbers show amino acid positions coded in the KUN NS1 and NS5 genes (8). probe, the cDNA fragment in the prM-E region that was used for Northern blot analysis (see Materials and Methods); a and b, the positions of the primers used in RT-PCR (Fig. 4A); HpaI, the location of the Hpa_I restriction site in the coding region introduced into FLd_GDD cDNA during construction (see reference 14). (B) Sindbis virus replicon constructs expressing KUN NS genes. The SR21IN vector was constructed from the noncytopathic DNA-based Sindbis virus replicon vector SINRep21 (2) as described in Materials and Methods. The KUN NS1 and NS5 genes and the KUN NS1-NS5 gene cassette were each cloned into the SR21IN vector as described in Materials and Methods to obtain the indicated SRns1, SRns5, and SRns1-5 constructs, respectively. XbaI-MluI, unique cloning sites; IresNeo, IRES of encephalomyelocarditis virus RNA followed by the NEO gene; RSV LTR, left terminal repeat of Rous sarcoma virus; 26S, Sindbis virus subgenomic promoter.
FIG. 2
Characterization of BHK cell lines stably expressing KUN NS genes from a Sindbis virus replicon vector. BHK cell lines expressing KUN NS genes were generated by transfection with corresponding recombinant Sindbis virus replicon plasmid DNAs followed by G418 selection as described in Materials and Methods. (A and C) IF analysis of SRns1BHK (A) and SRns1-5BHK (C) cells with anti-NS1 and anti-NS3 antibodies at passages 1 and 2, respectively. (B) WB analysis of SRns1BHK cells (passage 10) and repBHK cells (passage 10) with anti-NS1 antibodies. The SR21INBHK cells in panels A and C show results of IF analysis of the control cells expressing vector (SR21IN) RNA by using anti-NS1 and anti-NS3 antibodies, respectively. (D) WB analysis of SRns5BHK (passage 4), SRns1-5BHK (passage 4), and repBHK (passage 13) cells with anti-NS5 antibodies. Approximately 5 × 104 cells were boiled in the SDS-PAGE sample buffer without (B) or with (D) β-mercaptoethanol, and samples were electrophoresed in an SDS–12.5% (B) or –10% (D) polyacrylamide gel. Blotting and detection of expressed proteins were performed as described in Materials and Methods. (E) RIP analysis of SRns1-5BHK and vector control SR21INBHK cell lysates with anti-NS3 antibodies. The control KUN lane represents radiolabelled KUN-infected BHK cells, with dots indicating positions of labelled KUN proteins (from top to bottom) NS5, NS3, E, prM, NS2A, and NS2B/NS4A. SRns1-5BHKp13 and SR21INBHKp13 lanes represent results of RIP analysis of cells that were transfected with SRns1-5 and SR21IN (vector only) DNAs, respectively, and given 13 cell passages in medium containing 0.5 to 1 mg of G418 per ml. Confluent monolayers of cells in 60-mm culture dishes were preincubated with 6 μg of actinomycin D per ml in methionine-cysteine-free medium for 1 h at 37°C and then labelled for 6 h in the same medium supplemented with 50 μCi of [35S]methionine-cysteine for 6 h. The lysate was solubilized in RIP assay buffer (1% deoxycholate, 1% NP-40, 0.1% SDS, 0.1 M Tris-HCl [pH 7.5], 0.15 M NaCl) and centrifuged at 15,000 × g for 5 min to remove insoluble material, and a sample was radioimmunoprecipitated with anti-NS3 antibodies as described previously (33). The arrow shows the position of the NS3 protein.
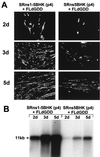
FIG. 3
Complementation of FLd_GDD_ RNA in SRns1-5BHK and SRns5BHK cells. (A) IF analysis of SRns5BHK (passage 4) and SRns1-5BHK (passage 4) cells at 2, 3, and 5 days after transfection with FLd_GDD_ RNA (14), using KUN anti-E antibodies. (B) Northern blot analysis of total RNA isolated from SRns5BHK (passage 4) and SRns1-5BHK (passage 4) cells at 2, 3, and 5 days after transfection with FLd_GDD_ RNA. The cDNA probe for the prM-E region was described in the legend for Fig. 1A. The arrow indicates the position of RNA at about 11 kb, determined relative to migration in the same gel of ethidium bromide-stained 1-kb Plus DNA ladder (Gibco BRL).
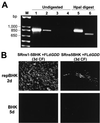
FIG. 4
Characterization of secreted complemented FLd_GDD_ viruses. (A) RT-PCR of complemented virus RNAs. KUN particles secreted from cells after complementation of the defective genomic RNA were treated with RNase A and DNase and immunoprecipitated with anti-E antibodies, and the virion RNA was extracted and used in RT-PCR analysis as described in Materials and Methods. Lane 1 represents an RT-PCR with KUN virion RNA purified as described previously (11). Lanes 2 to 4 represent the RT-PCRs of the RNA recovered from CFs after transfection of FLd_GDD_ RNA into SRns1-5BHK cells (lane 2), SRns5BHK cells (lane 3), and BHK cells (lane 4). Lanes 5 and 6 show an _Hpa_I digest of RT-PCR-amplified fragments from lanes 1 and 2, respectively. Lane M, molecular size markers. (B) IF analysis, using anti-E antibodies, of repBHK cells at 2 days and BHK cells at 5 days after infection with defective viruses recovered in 3-day CFs of FLd_GDD_-transfected SRns1-5BHK and SRns5BHK cells.
FIG. 5
Complementation of KUN NS1 mutant RNAs ns1/C10A and ns1/C11A in SRns1BHK and repBHK cells. (A) IF analysis of SRns1BHK, SR21INBHK, and repBHK cells with KUN anti-E antibodies at day 2 after transfection with ns1/C10A and ns1/C11A RNAs. (B) Northern blot analysis of total RNA isolated from SR21INBHK, SRns1BHK, and repBHK cells at 2 days after transfection with ns1/C10A and ns1/C11A RNAs. The probe (prM-E region) was the same as that in Fig. 3B. The arrow indicates the position of RNA at about 11 kb, determined as described in the legend for Fig. 3B.
FIG. 6
Characterization of complemented NS1 Cys− mutant viruses. (A) IF analysis, with anti-E antibodies, of SRns1BHK and SR21INBHK cells at 2 days after infection with 2-day CFs collected from SRns1BHK cells transfected with ns1/C10A or ns1/C11A RNAs. (B) IF analysis with anti-E antibodies of repBHK and BHK cells at 2 days (repBHK) and 5 days (BHK) after infection with 2-day CFs collected from repBHK cells transfected with ns1/C10A or ns1/C11A RNAs.
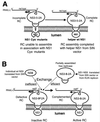
FIG. 7
Model showing how defective NS1 (NS1 Cys−) and NS5 with a deletion of the GDD polymerase motif (NS5d) in the RC can be complemented in trans by the wild-type (wt) NS1 and individual NS5 or NS1 to NS5, respectively, expressed from the Sindbis virus (SIN) replicon vector. (A) The RC assembles on the 3′UTR but is unable to associate correctly with the improperly folded NS1 Cys− mutant protein located in the lumen of the ER. However, this incomplete RC is able to associate with luminal wt NS1 supplied in trans from the Sindbis virus vector in SRns1BHK cells, thus completing the assembly of now functional RC. (B) A defective RC assembled with NS5d can be activated by an inefficient exchange with wt NS5 supplied independently in trans from the Sindbis virus vector in SRns5BHK cells. In contrast, a partially assembled wt KUN replicase supplied in trans by the Sindbis virus vector in SRns1-5BHK cells or by the KUN replicon in repBHK cells can readily exchange with the defective RC to form an active RC.
Similar articles
- trans-Complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells.
Khromykh AA, Kenney MT, Westaway EG. Khromykh AA, et al. J Virol. 1998 Sep;72(9):7270-9. doi: 10.1128/JVI.72.9.7270-7279.1998. J Virol. 1998. PMID: 9696822 Free PMC article. - trans-Complementation analysis of the flavivirus Kunjin ns5 gene reveals an essential role for translation of its N-terminal half in RNA replication.
Khromykh AA, Sedlak PL, Westaway EG. Khromykh AA, et al. J Virol. 1999 Nov;73(11):9247-55. doi: 10.1128/JVI.73.11.9247-9255.1999. J Virol. 1999. PMID: 10516033 Free PMC article. - cis- and trans-acting elements in flavivirus RNA replication.
Khromykh AA, Sedlak PL, Westaway EG. Khromykh AA, et al. J Virol. 2000 Apr;74(7):3253-63. doi: 10.1128/jvi.74.7.3253-3263.2000. J Virol. 2000. PMID: 10708442 Free PMC article. - Kunjin RNA replication and applications of Kunjin replicons.
Westaway EG, Mackenzie JM, Khromykh AA. Westaway EG, et al. Adv Virus Res. 2003;59:99-140. doi: 10.1016/s0065-3527(03)59004-2. Adv Virus Res. 2003. PMID: 14696328 Review. - Kunjin virus replicons: an RNA-based, non-cytopathic viral vector system for protein production, vaccine and gene therapy applications.
Pijlman GP, Suhrbier A, Khromykh AA. Pijlman GP, et al. Expert Opin Biol Ther. 2006 Feb;6(2):135-45. doi: 10.1517/14712598.6.2.135. Expert Opin Biol Ther. 2006. PMID: 16436039 Review.
Cited by
- Secretory pathways and multiple functions of nonstructural protein 1 in flavivirus infection.
Zhang S, He Y, Wu Z, Wang M, Jia R, Zhu D, Liu M, Zhao X, Yang Q, Wu Y, Zhang S, Huang J, Ou X, Gao Q, Sun D, Zhang L, Yu Y, Chen S, Cheng A. Zhang S, et al. Front Immunol. 2023 Jul 13;14:1205002. doi: 10.3389/fimmu.2023.1205002. eCollection 2023. Front Immunol. 2023. PMID: 37520540 Free PMC article. Review. - Identification of Key Residues in Dengue Virus NS1 Protein That Are Essential for Its Secretion.
Tan BEK, Beard MR, Eyre NS. Tan BEK, et al. Viruses. 2023 Apr 30;15(5):1102. doi: 10.3390/v15051102. Viruses. 2023. PMID: 37243188 Free PMC article. - In Vitro Infection Dynamics of Japanese Encephalitis Virus in Established Porcine Cell Lines.
Adetunji SA, Smolensky D, Mitzel DN, Owens JL, Chitko-McKown CG, Cernicchiaro N, Noronha LE. Adetunji SA, et al. Pathogens. 2021 Nov 12;10(11):1468. doi: 10.3390/pathogens10111468. Pathogens. 2021. PMID: 34832623 Free PMC article. - Roles of Non-Structural Protein 4A in Flavivirus Infection.
Klaitong P, Smith DR. Klaitong P, et al. Viruses. 2021 Oct 15;13(10):2077. doi: 10.3390/v13102077. Viruses. 2021. PMID: 34696510 Free PMC article. Review. - Molecular Insights into the Flavivirus Replication Complex.
van den Elsen K, Quek JP, Luo D. van den Elsen K, et al. Viruses. 2021 May 21;13(6):956. doi: 10.3390/v13060956. Viruses. 2021. PMID: 34064113 Free PMC article. Review.
References
- Adams S C, Brown A K, Sammels L M, Hartnett A C, Howard M J, Coelen R J, Mackenzie J S, Hall R A. Glycosylation and antigenic variation among Kunjin virus isolates. Virology. 1995;206:49–56. - PubMed
- Chu P W, Westaway E G. Replication strategy of Kunjin virus: evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology. 1985;140:68–79. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous