Box H and box ACA are nucleolar localization elements of U17 small nucleolar RNA - PubMed (original) (raw)
Box H and box ACA are nucleolar localization elements of U17 small nucleolar RNA
T S Lange et al. Mol Biol Cell. 1999 Nov.
Free PMC article
Abstract
The nucleolar localization elements (NoLEs) of U17 small nucleolar RNA (snoRNA), which is essential for rRNA processing and belongs to the box H/ACA snoRNA family, were analyzed by fluorescence microscopy. Injection of mutant U17 transcripts into Xenopus laevis oocyte nuclei revealed that deletion of stems 1, 2, and 4 of U17 snoRNA reduced but did not prevent nucleolar localization. The deletion of stem 3 had no adverse effect. Therefore, the hairpins of the hairpin-hinge-hairpin-tail structure formed by these stems are not absolutely critical for nucleolar localization of U17, nor are sequences within stems 1, 3, and 4, which may tether U17 to the rRNA precursor by base pairing. In contrast, box H and box ACA are major NoLEs; their combined substitution or deletion abolished nucleolar localization of U17 snoRNA. Mutation of just box H or just the box ACA region alone did not fully abolish the nucleolar localization of U17. This indicates that the NoLEs of the box H/ACA snoRNA family function differently from the bipartite NoLEs (conserved boxes C and D) of box C/D snoRNAs, where mutation of either box alone prevents nucleolar localization.
Figures
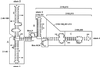
Figure 1
Structure and mutations of U17 snoRNA. The sequence and structure of Xenopus laevis U17 snoRNA (copy f), found in the sixth intron of the gene for ribosomal protein S7 (formerly S8), is from Cecconi et al. (1994), with sequence corrections at positions 33 (G instead of A), 91 and 161 (additional U). Shaded areas A and B in stems 1 and 3 of U17 are complementary to sequences in 18S rRNA (Cecconi et al., 1994). Shaded area C in stem 4 is complementary to the external transcribed spacer of pre-rRNA (Cecconi et al., 1994). The regions of U17 that were mutated in the present study are enclosed by lines. Box ACA, consisting of three nucleotides (enclosed by a dotted line), lies within the single-stranded 3′ end of the molecule. The sequences of the mutants of U17 designed for the present study are listed in the lower portion; nucleotides that are the same as in wild-type U14 snoRNA are shown by dots in the sequence alignment, and deletions are indicated by dashes. The double mutants Δbox H/Δbox ACA+, Δbox H/Δbox ACA, sub. box H/sub. box ACA+, Δ46–108/Δbox H, and Δ46–108/Δ152–213 are not shown, because they are simply the sum of the individual single mutations.
Figure 1
Structure and mutations of U17 snoRNA. The sequence and structure of Xenopus laevis U17 snoRNA (copy f), found in the sixth intron of the gene for ribosomal protein S7 (formerly S8), is from Cecconi et al. (1994), with sequence corrections at positions 33 (G instead of A), 91 and 161 (additional U). Shaded areas A and B in stems 1 and 3 of U17 are complementary to sequences in 18S rRNA (Cecconi et al., 1994). Shaded area C in stem 4 is complementary to the external transcribed spacer of pre-rRNA (Cecconi et al., 1994). The regions of U17 that were mutated in the present study are enclosed by lines. Box ACA, consisting of three nucleotides (enclosed by a dotted line), lies within the single-stranded 3′ end of the molecule. The sequences of the mutants of U17 designed for the present study are listed in the lower portion; nucleotides that are the same as in wild-type U14 snoRNA are shown by dots in the sequence alignment, and deletions are indicated by dashes. The double mutants Δbox H/Δbox ACA+, Δbox H/Δbox ACA, sub. box H/sub. box ACA+, Δ46–108/Δbox H, and Δ46–108/Δ152–213 are not shown, because they are simply the sum of the individual single mutations.
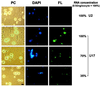
Figure 2
Nucleolar localization of wild-type U17 snoRNA injected into X. laevis oocytes. Fluorescein-labeled wild-type U17 snoRNA or spliceosomal U2 snRNA as a control was injected into the nuclei of X. laevis oocytes. After 1.5 h, nucleoli were prepared and analyzed by phase-contrast (PC) or fluorescence (FL) microscopy. The nucleolar rDNA is stained (DAPI, blue). Injection at an amount of 0.9 ng per oocyte (100%) resulted in a strong nucleolar labeling by U17 snoRNA but not by U2 snRNA (FL, green). After dilution of U17, even 35% of this amount yields detectable nucleolar labeling 1.5 h after injection. Oocyte nucleoli vary in size (Wu and Gall, 1997) and can fuse into multinucleolar clusters (Shah et al., 1996). A lampbrush chromosome is visible by DAPI staining (see PC and DAPI panels for 70% of U17 injected). Bar, 10 μm.
Figure 3
Nucleolar localization of U17 snoRNA after deletion of sequences with stem structures and rRNA binding sites. Fluorescein-labeled U17 snoRNA was injected into the nuclei of X. laevis oocytes at an amount of 0.9 ng per oocyte. After 1.5 h, nucleolar preparations were analyzed by phase-contrast (PC) or fluorescence (FL) microscopy; the nucleolar rDNA was stained (DAPI, blue). U17 snoRNA carrying a deletion of stem 3 (Δ118–151) localized as well to nucleoli as the wild-type molecule (FL, green). U17 deleted in stem 1 (Δ1–45) localized strongly to nucleoli, and deletions of stem 2 (Δ46–108), stem 4 (Δ152–213), or a combination of both (Δ46–108/Δ152–213) showed significantly less but not abolished localization. Dissection of stem 4 (Δ169–200 or Δ152–168,201–213) did not reveal any major site of importance for nucleolar localization. The deletion of the entire structure of stems 3 and 4 between the single-stranded regions of conserved boxes H and ACA (Δ118–213) reduced but did not completely abolish nucleolar localization. Bar, 10 μm.
Figure 4
Role of evolutionarily conserved box H and box ACA in nucleolar localization of U17 snoRNA. U17 snoRNA carrying a substitution (sub. box H) or deletion (Δbox H) of just conserved box H alone retained the ability to localize to nucleoli, resulting in moderate to strong nucleolar labeling. Nucleolar localization signals with mutants substituted in box ACA, deleted in the ACA region (Δbox ACA+), or substituted in the ACA region (sub. box ACA+) were highly variable. The double mutants of box H and the box ACA region, being either depleted (Δbox H/Δbox ACA+) or substituted (sub. box H/sub. box ACA+) in both sequences, were not capable of localizing to nucleoli. Nucleolar localization was also abolished when the deletion of box H was coupled with the deletion of just box ACA itself (Δbox H/Δbox ACA). A combination of the deletion of stem 2 and box H (Δ46–108/Δbox H) did not enhance the effect of a stem 2 deletion alone (see Figure 3), and localization was still apparent. Bar, 10 μm.
Figure 5
Stability of wild-type and mutated U17 snoRNA. 32P-labeled U17 snoRNA (mutants or wild-type) were injected into oocyte nuclei, and the RNAs were isolated and analyzed by gel 8% polyacrylamide, 8 M urea gel electrophoresis. The upper panel shows controls (sample recovery immediately after injection, 0 h), the middle panel shows short-term stability at 1.5 h (the time when localization assays were carried out), and the lower panel shows long-term stability (left gel at 15 h, right gel at 16 h) of U17 snoRNA. 32P-labeled U14 snoRNA (lower band) was coinjected as an internal control to normalize for any differences in injection or recovery of the samples. The relative stability was calculated as [U17/U14 after incubation]/[U17/U14 at 0 h]. The ratio of U17/U14 shows that all the mutants are stable at the 1.5-h time point used for analysis of nucleolar localization (middle panel).
Similar articles
- Comparative structure analysis of vertebrate U17 small nucleolar RNA (snoRNA).
Cervelli M, Cecconi F, Giorgi M, Annesi F, Oliverio M, Mariottini P. Cervelli M, et al. J Mol Evol. 2002 Feb;54(2):166-79. doi: 10.1007/s00239-001-0065-2. J Mol Evol. 2002. PMID: 11821910 - Nucleolar localization signals of box H/ACA small nucleolar RNAs.
Narayanan A, Lukowiak A, Jády BE, Dragon F, Kiss T, Terns RM, Terns MP. Narayanan A, et al. EMBO J. 1999 Sep 15;18(18):5120-30. doi: 10.1093/emboj/18.18.5120. EMBO J. 1999. PMID: 10487763 Free PMC article. - Nucleolar localization elements in U8 snoRNA differ from sequences required for rRNA processing.
Lange TS, Borovjagin AV, Gerbi SA. Lange TS, et al. RNA. 1998 Jul;4(7):789-800. doi: 10.1017/s1355838298980438. RNA. 1998. PMID: 9671052 Free PMC article. - The vertebrate E1/U17 small nucleolar ribonucleoprotein particle.
Eliceiri GL. Eliceiri GL. J Cell Biochem. 2006 Jun 1;98(3):486-95. doi: 10.1002/jcb.20821. J Cell Biochem. 2006. PMID: 16475166 Review. - RNA structure and function in C/D and H/ACA s(no)RNPs.
Henras AK, Dez C, Henry Y. Henras AK, et al. Curr Opin Struct Biol. 2004 Jun;14(3):335-43. doi: 10.1016/j.sbi.2004.05.006. Curr Opin Struct Biol. 2004. PMID: 15193314 Review.
Cited by
- The path from student to mentor and from chromosomes to replication to genomics.
Gerbi SA. Gerbi SA. Mol Biol Cell. 2016 Nov 1;27(21):3194-3196. doi: 10.1091/mbc.E16-07-0493. Mol Biol Cell. 2016. PMID: 27799493 Free PMC article. - RNA pseudouridylation: new insights into an old modification.
Ge J, Yu YT. Ge J, et al. Trends Biochem Sci. 2013 Apr;38(4):210-8. doi: 10.1016/j.tibs.2013.01.002. Epub 2013 Feb 4. Trends Biochem Sci. 2013. PMID: 23391857 Free PMC article. Review. - Insight into the Protein Components of the Box H/ACA RNP.
Karijolich J, Yu YT. Karijolich J, et al. Curr Proteomics. 2008 Jul 1;5(2):129-137. doi: 10.2174/157016408784911936. Curr Proteomics. 2008. PMID: 19829749 Free PMC article. - Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10.
Pogacić V, Dragon F, Filipowicz W. Pogacić V, et al. Mol Cell Biol. 2000 Dec;20(23):9028-40. doi: 10.1128/MCB.20.23.9028-9040.2000. Mol Cell Biol. 2000. PMID: 11074001 Free PMC article. - Transient nucleolar localization Of U6 small nuclear RNA in Xenopus Laevis oocytes.
Lange TS, Gerbi SA. Lange TS, et al. Mol Biol Cell. 2000 Jul;11(7):2419-28. doi: 10.1091/mbc.11.7.2419. Mol Biol Cell. 2000. PMID: 10888678 Free PMC article.
References
- Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different Box elements with related functions. Cell. 1996;86:823–834. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources