Recognition of yeast mRNAs as "nonsense containing" leads to both inhibition of mRNA translation and mRNA degradation: implications for the control of mRNA decapping - PubMed (original) (raw)
Recognition of yeast mRNAs as "nonsense containing" leads to both inhibition of mRNA translation and mRNA degradation: implications for the control of mRNA decapping
D Muhlrad et al. Mol Biol Cell. 1999 Nov.
Free PMC article
Abstract
A critical step in the degradation of many eukaryotic mRNAs is a decapping reaction that exposes the transcript to 5' to 3' exonucleolytic degradation. The dual role of the cap structure as a target of mRNA degradation and as the site of assembly of translation initiation factors has led to the hypothesis that the rate of decapping would be specified by the status of the cap binding complex. This model makes the prediction that signals that promote mRNA decapping should also alter translation. To test this hypothesis, we examined the decapping triggered by premature termination codons to determine whether there is a down-regulation of translation when mRNAs were recognized as "nonsense containing." We constructed an mRNA containing a premature stop codon in which we could measure the levels of both the mRNA and the polypeptide encoded upstream of the premature stop codon. Using this system, we analyzed the effects of premature stop codons on the levels of protein being produced per mRNA. In addition, by using alterations either in cis or in trans that inactivate different steps in the recognition and degradation of nonsense-containing mRNAs, we demonstrated that the recognition of a nonsense codon led to a decrease in the translational efficiency of the mRNA. These observations argue that the signal from a premature termination codon impinges on the translation machinery and suggest that decapping is a consequence of the change in translational status of the mRNA.
Figures
Figure 1
Northern analysis of the PGK1(cup1) containing transcripts. Diagrams on the left represent the mRNAs: PGK1(cup1)pG, PGK1(cup1)UAApG, and PGK1(cup1)UAApG(ΔDSE). They show the location of the insertions of the Flag tag, the CUP1 open reading frame, both the premature and normal stop codons, and a poly(G) tag sequence. The region of the DSE deletion is designated with two dashed lines between the diagrams and is depicted as Δ. Strain names are labeled to the right of the construct cartoons; WT, wild-type. Northern blots showing decay after transcriptional repression are shown for each construct in the respective strains. Above each panel is the time in minutes after repression of transcription. Average half-life values from at least two experiments are given to the right of each Northern blot.
Figure 2
Relative mRNA levels of the PGK1(cup1)UAApG and the PGK1(cup1)UAApG(ΔDSE) transcripts. The upper Northern blot shows analysis of the PGK1(cup1)UAApG (left four lanes) and the PGK1(cup1)UAApG(ΔDSE) (right four lanes) transcripts. Yeast strains are designated above each lane as follows: wild type (WT), dcp1Δ (D), upf1Δ (U), and dcp1Δupf1Δ (D/U). The relative percent mRNA levels and error values for each mRNA species from five or more experiments are shown below each lane, with the PGK1(cup1)UAApG mRNA in the wild-type strain defined as 100%. The lower panel shows the same Northern blot probed for the 7S ribosomal RNA, which was used to standardize the lanes.
Figure 3
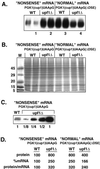
Relative protein levels of the PGK1(cup1)UAApG and the PGK1(cup1)UAApG(ΔDSE) transcripts in wild-type and upf1Δ strains. (A) Western analysis using the FLAG antibody to measure protein levels produced from the PGK1(cup1)UAApG and PGK1(cup1)UAApG(ΔDSE) constructs, labeled at the top. Strain names are given above each lane set, which consist of two independent experiments run side by side. One lane of each set is numbered left to right as reference for the text. (B) Stained protein gel containing the identical samples to A. (C) Dilution comparison Western analysis of the PGK1(cup1)UAApG containing wild-type (WT) and upf1Δ strains. Dilution factors are given below each lane. (D) The percent relative mRNA and protein levels as well as protein per mRNA ratios are given for each sample type. The level of the protein produced from the nonsense mRNA PGK1(cup1)UAApG in wild-type cells is set at 100%. Values are averages based on at least five independent experiments.
Figure 4
Copper phenotypes of the PGK1(cup1)UAApG and the PGK1(cup1)UAApG(ΔDSE) transcripts in wild-type and deletion strains. The top portion shows yeast colonies containing the PGK1(cup1)UAApG construct from four different strains designated on the left. From top to bottom: wild-type (WT), upf1Δ, dcp1Δ, and the double deletion dcp1Δupf1Δ. Colonies were grown for 3 d on selective media containing galactose and increasing millimolar levels of copper designated above each column. The lower panels are identical to above, except that these strains contain the PGK1(cup1)UAApG(ΔDSE) construct.
Figure 5
Relative protein levels of the PGK1(cup1)UAApG and the PGK1(cup1)UAApG(ΔDSE) transcripts in dcp1Δ and dcp1Δupf1Δ strains. (A) Western analysis of PGK1(cup1)UAApG and PGK1(cup1)UAApG(ΔDSE) constructs in the wild-type (WT), dcp1Δ, and dcp1Δupf1Δ strains. Strain names and constructs are given above the Western blots, and lanes are labeled below as a reference for the text. FLAG antibodies were used to probe the Western blots. (B) Values for protein levels, percent mRNA levels, and ratios of protein per mRNA are shown for each lane corresponding to the Western blots in A. These values are from at least five independent experiments. To allow comparisons, we have set the levels of the protein produced from the nonsense mRNA PGK1(cup1)UAApG in dcp1Δ cells at 100%.
Similar articles
- Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs.
He F, Jacobson A. He F, et al. Mol Cell Biol. 2001 Mar;21(5):1515-30. doi: 10.1128/MCB.21.5.1515-1530.2001. Mol Cell Biol. 2001. PMID: 11238889 Free PMC article. - Aberrant mRNAs with extended 3' UTRs are substrates for rapid degradation by mRNA surveillance.
Muhlrad D, Parker R. Muhlrad D, et al. RNA. 1999 Oct;5(10):1299-307. doi: 10.1017/s1355838299990829. RNA. 1999. PMID: 10573121 Free PMC article. - mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E.
Schwartz DC, Parker R. Schwartz DC, et al. Mol Cell Biol. 2000 Nov;20(21):7933-42. doi: 10.1128/MCB.20.21.7933-7942.2000. Mol Cell Biol. 2000. PMID: 11027264 Free PMC article. - Eukaryotic mRNA decapping.
Coller J, Parker R. Coller J, et al. Annu Rev Biochem. 2004;73:861-90. doi: 10.1146/annurev.biochem.73.011303.074032. Annu Rev Biochem. 2004. PMID: 15189161 Review. - Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae.
Tucker M, Parker R. Tucker M, et al. Annu Rev Biochem. 2000;69:571-95. doi: 10.1146/annurev.biochem.69.1.571. Annu Rev Biochem. 2000. PMID: 10966469 Review.
Cited by
- Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs.
He F, Jacobson A. He F, et al. Mol Cell Biol. 2001 Mar;21(5):1515-30. doi: 10.1128/MCB.21.5.1515-1530.2001. Mol Cell Biol. 2001. PMID: 11238889 Free PMC article. - How do trypanosomes change gene expression in response to the environment?
Schwede A, Kramer S, Carrington M. Schwede A, et al. Protoplasma. 2012 Apr;249(2):223-38. doi: 10.1007/s00709-011-0282-5. Epub 2011 May 20. Protoplasma. 2012. PMID: 21594757 Free PMC article. Review. - When a ribosome encounters a premature termination codon.
Hwang J, Kim YK. Hwang J, et al. BMB Rep. 2013 Jan;46(1):9-16. doi: 10.5483/bmbrep.2013.46.1.002. BMB Rep. 2013. PMID: 23351378 Free PMC article. Review. - Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes.
Mendell JT, Medghalchi SM, Lake RG, Noensie EN, Dietz HC. Mendell JT, et al. Mol Cell Biol. 2000 Dec;20(23):8944-57. doi: 10.1128/MCB.20.23.8944-8957.2000. Mol Cell Biol. 2000. PMID: 11073994 Free PMC article. - Human MARF1 is an endoribonuclease that interacts with the DCP1:2 decapping complex and degrades target mRNAs.
Nishimura T, Fakim H, Brandmann T, Youn JY, Gingras AC, Jinek M, Fabian MR. Nishimura T, et al. Nucleic Acids Res. 2018 Dec 14;46(22):12008-12021. doi: 10.1093/nar/gky1011. Nucleic Acids Res. 2018. PMID: 30364987 Free PMC article.
References
- Beelman CA, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. - PubMed
- Beelman CA, Stevens A, Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous