Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae - PubMed (original) (raw)
Review
Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae
D Kressler et al. Mol Cell Biol. 1999 Dec.
No abstract available
Figures
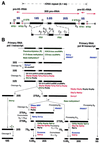
FIG. 1
Pre-rRNA processing in S. cerevisiae. (A) Structure of an rDNA repeat unit. Each unit contains a large operon encoding the 18S, 5.8S, and 25S rRNAs, which is transcribed by RNA pol I (pr, promoter; tr, terminator; eh, enhancer), and an RNA pol III-transcribed 5S rRNA gene. Mature sequences of the RNA pol I transcript are separated by ITS1 and ITS2 and flanked by a 5′ ETS and a 3′ ETS. The 5S rRNA gene is located between two nontranscribed spacers (NTS1 and NTS2). Black boxes represent mature rRNAs, white thin bars represent the transcribed spacers, and lines represent the nontranscribed spacers. Processing sites are also indicated. (B) Pre-rRNA processing pathways. The primary RNA pol I transcript is processed at its 3′ end to yield the 35S pre-rRNA, which is the longest detectable pre-rRNA. The 35S pre-rRNA is first processed at sites A0, A1, and A2, which results in the separation of the pre-rRNAs destined for the small and large ribosomal subunits. The 20S pre-rRNA is matured by endonucleolytic cleavage at site D. The 27SA2 precursor is processed by two alternative pathways. In the major pathway, about 85% of 27SA2 pre-rRNA is cleaved at site A3 and then 5′→3′ exonucleolytically digested to site B1S. In the minor pathway, about 15% of the 27SA2 molecules are processed at site B1L. While processing at site B1 is completed, the 3′ end of mature 25S rRNA is generated by processing at site B2. The subsequent ITS2 processing of both 27SB species appears to be the same. Cleavage at sites C1 and C2 releases the mature 25S rRNA and the 7S pre-rRNA. The latter undergoes 3′→5′ exonucleolytic digestion to the 3′ end of the mature 5.8S rRNA. The pre-5S rRNA is processed at its 3′ end to generate the mature 5S rRNA. Pseudouridylation and 2′-_O_-ribose methylation take place on the primary transcript. Note that except for Dim1p, the timing of base methylation and the protein _trans_-acting factors involved therein have not yet been uncovered. Factors involved in pre-rRNA processing and modification are shown. Colors were assigned to each class of protein _trans_-acting factors as follows: blue, exo- and endonucleases; red, putative ATP-dependent RNA helicases; green, components involved in pre-rRNA modification; black, others. See the text for further details.
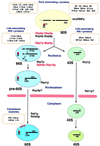
FIG. 2
Ribosomal subunit assembly pathway in S. cerevisiae. In the nucleolus, the 35S pre-rRNA associates with many early-associating 40S and 60S r-proteins to form a 90S preribosome. The 5S rRNA forms a stable RNP with Rpl5p (L5, red dot). The 5S rRNA is probably already present in 90S preribosomes; therefore, Rpl5p may belong to the group of early-assembling 60S r-proteins. From the 90S particle, 66S and 43S preribosomes containing the 27S and 20S pre-rRNAs, respectively, are formed. The 66S and the 43S preribosomes contain the late-associating r-proteins. Preribosomes are most likely actively exported through nuclear pores. It has been demonstrated that export of 60S ribosomal subunits requires the nuclear or cytoplasmic Ran cycle and distinct nucleoporins (not shown; for details, see reference 70). It is believed that the 66S particle remains in the nucleus until the 7S pre-rRNA is processed to the mature 5.8S rRNA, while the 43S preribosome is rapidly exported to the cytoplasm, where the final maturation step in the synthesis of the 18S rRNA takes place. Assembly of 60S ribosomal subunits is also completed in the cytoplasm by the final incorporation of some r-proteins. At least three of them (also depicted as red dots), including the P2B and Rpl10p/Qsr1p (L10) r-proteins, can exchange on mature 60S ribosomal subunits in vivo. Preribosomes are drawn as balloons, and the nuclear envelope is depicted as pink rods. Protein _trans_-acting factors with likely involvement in the assembly of ribosomal subunits are colored as follows: red, putative ATP-dependent RNA helicases; black, others. Only those r-proteins for which an indication about the timing of their association with preribosomal particles is known are shown. See the text for further details.
Similar articles
- Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing.
Sloan KE, Mattijssen S, Lebaron S, Tollervey D, Pruijn GJ, Watkins NJ. Sloan KE, et al. J Cell Biol. 2013 Mar 4;200(5):577-88. doi: 10.1083/jcb.201207131. Epub 2013 Feb 25. J Cell Biol. 2013. PMID: 23439679 Free PMC article. - Ribosome synthesis in Saccharomyces cerevisiae.
Venema J, Tollervey D. Venema J, et al. Annu Rev Genet. 1999;33:261-311. doi: 10.1146/annurev.genet.33.1.261. Annu Rev Genet. 1999. PMID: 10690410 Review. - The role of small nucleolar ribonucleoproteins in ribosome synthesis.
Tollervey D, Hurt EC. Tollervey D, et al. Mol Biol Rep. 1990;14(2-3):103-6. doi: 10.1007/BF00360433. Mol Biol Rep. 1990. PMID: 2141891 Review. No abstract available. - Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae.
Emery B, de la Cruz J, Rocak S, Deloche O, Linder P. Emery B, et al. Mol Microbiol. 2004 Apr;52(1):141-58. doi: 10.1111/j.1365-2958.2003.03973.x. Mol Microbiol. 2004. PMID: 15049817 - Rat1p and Rai1p function with the nuclear exosome in the processing and degradation of rRNA precursors.
Fang F, Phillips S, Butler JS. Fang F, et al. RNA. 2005 Oct;11(10):1571-8. doi: 10.1261/rna.2900205. Epub 2005 Aug 30. RNA. 2005. PMID: 16131592 Free PMC article.
Cited by
- Synergistic defect in 60S ribosomal subunit assembly caused by a mutation of Rrs1p, a ribosomal protein L11-binding protein, and 3'-extension of 5S rRNA in Saccharomyces cerevisiae.
Nariai M, Tanaka T, Okada T, Shirai C, Horigome C, Mizuta K. Nariai M, et al. Nucleic Acids Res. 2005 Aug 12;33(14):4553-62. doi: 10.1093/nar/gki772. Print 2005. Nucleic Acids Res. 2005. PMID: 16100378 Free PMC article. - The Noc-domain containing C-terminus of Noc4p mediates both formation of the Noc4p-Nop14p submodule and its incorporation into the SSU processome.
Kühn H, Hierlmeier T, Merl J, Jakob S, Aguissa-Touré AH, Milkereit P, Tschochner H. Kühn H, et al. PLoS One. 2009 Dec 18;4(12):e8370. doi: 10.1371/journal.pone.0008370. PLoS One. 2009. PMID: 20019888 Free PMC article. - Dbp9p, a putative ATP-dependent RNA helicase involved in 60S-ribosomal-subunit biogenesis, functionally interacts with Dbp6p.
Daugeron MC, Kressler D, Linder P. Daugeron MC, et al. RNA. 2001 Sep;7(9):1317-34. doi: 10.1017/s1355838201010640. RNA. 2001. PMID: 11565753 Free PMC article. - NVL2 is a nucleolar AAA-ATPase that interacts with ribosomal protein L5 through its nucleolar localization sequence.
Nagahama M, Hara Y, Seki A, Yamazoe T, Kawate Y, Shinohara T, Hatsuzawa K, Tani K, Tagaya M. Nagahama M, et al. Mol Biol Cell. 2004 Dec;15(12):5712-23. doi: 10.1091/mbc.e04-08-0692. Epub 2004 Oct 6. Mol Biol Cell. 2004. PMID: 15469983 Free PMC article. - Nascent 60S ribosomal subunits enter the free pool bound by Nmd3p.
Ho JH, Kallstrom G, Johnson AW. Ho JH, et al. RNA. 2000 Nov;6(11):1625-34. doi: 10.1017/s1355838200001291. RNA. 2000. PMID: 11105761 Free PMC article.
References
- Abou Elela S, Igel H, Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. - PubMed
- Allende M L, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development. Genes Dev. 1996;10:3141–3155. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases