Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex - PubMed (original) (raw)
Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex
S Ceman et al. Mol Cell Biol. 1999 Dec.
Abstract
The loss of FMR1 expression due to trinucleotide repeat expansion leads to fragile X syndrome, a cause of mental retardation. The encoded protein, FMRP, is a member of a gene family that also contains the fragile X-related proteins, FXR1P and FXR2P. FMRP has been shown to be a nucleocytoplasmic shuttling protein that selectively binds a subset of mRNAs, forms messenger ribonucleoprotein (mRNP) complexes, and associates with translating ribosomes. Here we describe a cell culture system from which we can isolate epitope-tagged FMRP along with mRNA, including its own message, and at least six other proteins. We identify two of these proteins as FXR1P and FXR2P by using specific antisera and identify a third protein as nucleolin by using mass spectrometry. The presence of nucleolin is confirmed by both reactivity with a specific antiserum as well as reverse coimmunoprecipitation where antinucleolin antiserum immunoprecipitates endogenous FMRP from both cultured cells and mouse brain. The identification of nucleolin, a known component of other mRNPs, adds a new dimension to the analysis of FMRP function, and the approach described should also allow the identification of the remaining unknown proteins of this FMRP-associated mRNP as well as the other bound mRNAs.
Figures
FIG. 1
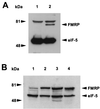
Transfected murine L-M(TK−) cells express epitope-tagged FMRP at levels comparable to that observed in a transformed B-cell line. (A) Approximately 2 × 105 cell equivalents from L-M(TK−) cells expressing either vector only (lane 1) or Flag-FMRP were loaded (lane 2). Positions of molecular weight markers are shown on the left; positions of Flag-FMRP and eIF-5 are indicated by arrowheads on the right. The cytoplasmic proteins were resolved on a 7.5% gel, transferred to nitrocellulose, and probed simultaneously with anti-Flag monoclonal antibody M2 and with a monoclonal antibody that recognizes eIF-5 to show that equal amounts of cytoplasmic lysate were loaded. The upper band (>81 kDa) present in both lanes is detected by this particular goat anti-mouse HRP-conjugated antibody alone (data not shown). The secondary antibodies used for panel A are different from those used for panel B. (B) Approximately 5 × 105 cell equivalents of cytoplasmic lysates from untransfected L-M(TK−) cells (lane 1), two independently derived clones expressing Flag-tagged FMRP (lanes 2 and 3), and an Epstein-Barr virus-transformed B-cell line (J1; lane 4) were loaded per lane of a 12% gel. After transfer, the blot was probed simultaneously with anti-FMRP monoclonal antibody 1FM.1AC.484A.1 and eIF-5 to show equal loading.
FIG. 2
FXR1P and FXR2P assemble with Flag-FMRP in transfected L-M(TK−) cells to form an mRNP particle that binds mRNA. (A) Cytoplasmic lysates from approximately 5 × 105 L-M(TK−) cells expressing vector alone and expressing Flag-FMRP were loaded into lanes 1 and 2, respectively; lanes 3 to 5 contain Flag peptide elutions from the anti-Flag antibody M2 alone (lane 3) or from immunoprecipitations of 107 cells expressing the vector -only (lane 4) or Flag-FMRP (lane 5). The immunoprecipitated FMRP in lane 5 appears to run slower than the FMRP detected in the cytoplasmic lysates, probably because there is much less protein in the lanes containing the peptide elutions than in the lanes containing cytoplasmic lysates. The gel was blotted and sequentially probed with a monoclonal antibody recognizing FMRP (A), then with both anti-FMRP and anti-FXR2P antibodies (B), and finally with anti-FMRP, anti-FXR2P and anti-FXR1P antibodies (C). Lanes 1 and 2 in panel C are shown as separate because they are a lighter exposure of the same blot. Positions of the molecular weight standards are shown on the left, and positions of the proteins are shown on the right. The long and short isoforms of FXR1P are indicated by lines. (D) mRNA was purified from L-M(TK−) cells expressing either the vector only (lane 1) or Flag-FMRP (lane 2) as described in Materials and Methods. The mRNA was recovered, and the polyadenylated species were labeled by priming with oligo(dT) and synthesizing first-strand cDNA with reverse transcriptase. (E) MRNA obtained from immunoprecipitations of L-M(TK−) cells expressing either Flag-FMRP (lanes 1 and 2) or vector alone (lanes 3 and 4) or from mouse brain (lanes 5 and 6) was reverse transcribed with an oligo(dT) primer in either the presence (lanes 2, 4, and 6) or the absence (lanes 1, 3, and 5) of reverse transcriptase. A fraction of each reaction mixture was then added to a PCR mixture with mouse FMR1 primers. The PCR products were resolved on an agarose gel and stained with ethidium bromide.
FIG. 3
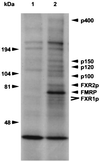
Novel proteins in addition to FXR1P and FXR2P assemble with Flag-FMRP in L-M(TK−) cells. L-M(TK−) cells transfected with either the eukaryotic expression vector alone (lane 1) or with Flag-FMRP (lane 2) were labeled overnight with [3H]leucine and then immunoprecipitated with matrix coupled to anti-Flag antibody M2. After extensive washing, the Flag matrix was eluted with Flag peptide and the proteins were resolved on a 5 to 20% gradient gel. Migration of molecular weight markers is indicated on the left. Known proteins are indicated on the right, and the new proteins are indicated by their molecular sizes. Both the long and short isoforms of FXR1P are indicated, and FMRP is highlighted.
FIG. 4
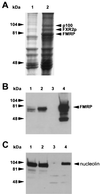
Nucleolin coimmunoprecipitates with Flag-FMRP. (A) Scanned image of the Coomassie brilliant blue-stained gel from which p100 was harvested. Lanes 1 and 2 are the Flag peptide elutions from the large-scale purifications of L-M(TK−) cells expressing vector alone and Flag-FMRP, respectively. The proteins were resolved on 7.5% minigels. The position of FMRP was determined by Western blotting of a gel run in parallel (B). The position of FXR2 was determined by both Western blotting and mass analysis (data not shown). The p100 band was cut out and analyzed as described in the text. (B) FMRP Western analysis of the large-scale purification. Lanes 1 and 2 contain a fraction of the pooled lysates from L-M(TK−) cells expressing the vector and Flag-FMRP before immunoprecipitation, respectively; lanes 3 and 4 show 7.5% of the peptide elutions from each of the large-scale purifications. (C) A rabbit antiserum derived against murine nucleolin was used to reprobe the Western blot shown in panel B. Positions of the molecular weight markers are shown on the left; the position of nucleolin is shown on the right.
FIG. 5
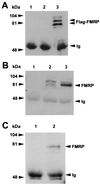
FMRP coimmunoprecipitates with nucleolin. (A) Lane 1 contains the antinucleolin antibody alone; lanes 2 and 3 show antinucleolin immunoprecipitation of cytoplasmic lysates from 107 L-M(TK−) cells expressing the vector alone and from 107 L-M(TK−) cells expressing Flag-FMRP, respectively. The proteins were resolved on a 7.5% gel, blotted to nitrocellulose, and probed with anti-Flag antibody M2. Positions of the molecular weight markers are shown on the left, and positions of FMRP and the heavy chain of the antinucleolin antibody (immunoglobulin [Ig]) are shown on the right. (B) An experiment similar to that shown in panel A except that transferred proteins were probed with the anti-FMRP antibody. Lanes 1 to 3 are as described above: the antinucleolin antibody alone, an immunoprecipitation of vector-only-containing L-M(TK−) cells, and an immunoprecipitation of Flag-FMRP-expressing L-M(TK−) cells with the antinucleolin antibody. In lane 2, the endogenous murine FMRP is immunoprecipitated with nucleolin in addition to the Flag-tagged FMRP observed in lane 3. Positions of the heavy chain of the antinucleolin antibody, which reacts with the second-step goat anti-mouse HRP conjugate, and Flag-FMRP are indicated on the right. (C) Total brain homogenates were prepared from either FMR1 knockout mice (lane 1) or their wild-type, FMRP-positive littermates (lane 2). The cytoplasmic lysates were immunoprecipitated with the antinucleolin antibody, washed extensively, and boiled. The samples were resolved on a 7.5% gel, transferred to nitrocellulose, and then probed with a monoclonal antibody that recognizes FMRP. Positions of FMRP and the heavy chain of the antinucleolin antibody are shown on the right.
FIG. 6
Treatment with RNase does not affect the association of nucleolin with FMRP. (A) Lanes 1 and 2 contain cytoplasmic lysates from 2.5 × 105 L-M(TK−) cells expressing the vector alone and from L-M(TK−) cells expressing Flag-FMRP, respectively; lanes 3 and 4 contain mock-treated, anti-Flag antibody immunoprecipitations from L-M(TK−) cells expressing the vector alone and from L-M(TK−) cells expressing Flag-FMRP, respectively; lanes 5 and 6 contain RNase-treated anti-Flag antibody immunoprecipitations from L-M(TK−) cells expressing the vector alone and from L-M(TK−) cells expressing Flag-FMRP, respectively. The proteins were resolved on a 7.5% gel, blotted to nitrocellulose, and probed with an antinucleolin antibody. Positions of molecular weight markers are shown on the left, and the position of nucleolin is shown on the right. (B) An experiment similar to that shown in panel A except that the transferred proteins were probed with an antibody to hnRNP A1. Lanes 1 and 2 are as described above except that lysates from 5 × 104 cells were loaded. Lanes 3 and 4 contain mock-treated anti-Flag antibody immunoprecipitations from L-M(TK−) cells expressing the vector alone and from L-M(TK−) cells expressing Flag-FMRP; lanes 5 and 6 contain RNase-treated anti-Flag antibody immunoprecipitations from L-M(TK−) cells expressing the vector alone and from L-M(TK−) cells expressing Flag-FMRP. The proteins were resolved on a 10% gel, blotted to nitrocellulose, and probed with an antibody that recognizes hnRNP A1. Positions of molecular weight markers are shown on the left, and the position of hnRNP A1 is shown on the right.
Similar articles
- Oligomerization properties of fragile-X mental-retardation protein (FMRP) and the fragile-X-related proteins FXR1P and FXR2P.
Tamanini F, Van Unen L, Bakker C, Sacchi N, Galjaard H, Oostra BA, Hoogeveen AT. Tamanini F, et al. Biochem J. 1999 Nov 1;343 Pt 3(Pt 3):517-23. Biochem J. 1999. PMID: 10527928 Free PMC article. - Purified recombinant Fmrp exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein.
Brown V, Small K, Lakkis L, Feng Y, Gunter C, Wilkinson KD, Warren ST. Brown V, et al. J Biol Chem. 1998 Jun 19;273(25):15521-7. doi: 10.1074/jbc.273.25.15521. J Biol Chem. 1998. PMID: 9624140 - Identification of mouse YB1/p50 as a component of the FMRP-associated mRNP particle.
Ceman S, Nelson R, Warren ST. Ceman S, et al. Biochem Biophys Res Commun. 2000 Dec 29;279(3):904-8. doi: 10.1006/bbrc.2000.4035. Biochem Biophys Res Commun. 2000. PMID: 11162447 - Biology of the fragile X mental retardation protein, an RNA-binding protein.
Khandjian EW. Khandjian EW. Biochem Cell Biol. 1999;77(4):331-42. Biochem Cell Biol. 1999. PMID: 10546896 Review. - The RNA binding protein FMRP: new connections and missing links.
Schaeffer C, Beaulande M, Ehresmann C, Ehresmann B, Moine H. Schaeffer C, et al. Biol Cell. 2003 May-Jun;95(3-4):221-8. doi: 10.1016/s0248-4900(03)00037-6. Biol Cell. 2003. PMID: 12867085 Review.
Cited by
- The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif.
Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. Schaeffer C, et al. EMBO J. 2001 Sep 3;20(17):4803-13. doi: 10.1093/emboj/20.17.4803. EMBO J. 2001. PMID: 11532944 Free PMC article. - Nervous translation, do you get the message? A review of mRNPs, mRNA-protein interactions and translational control within cells of the nervous system.
Smith R, Rathod RJ, Rajkumar S, Kennedy D. Smith R, et al. Cell Mol Life Sci. 2014 Oct;71(20):3917-37. doi: 10.1007/s00018-014-1660-x. Epub 2014 Jun 22. Cell Mol Life Sci. 2014. PMID: 24952431 Free PMC article. Review. - Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation.
Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Valinezhad Orang A, et al. Int J Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. Epub 2014 Aug 10. Int J Genomics. 2014. PMID: 25180174 Free PMC article. Review. - Fmr1 KO and fenobam treatment differentially impact distinct synapse populations of mouse neocortex.
Wang GX, Smith SJ, Mourrain P. Wang GX, et al. Neuron. 2014 Dec 17;84(6):1273-86. doi: 10.1016/j.neuron.2014.11.016. Neuron. 2014. PMID: 25521380 Free PMC article. - FMRP targets distinct mRNA sequence elements to regulate protein expression.
Ascano M Jr, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, Williams Z, Ohler U, Tuschl T. Ascano M Jr, et al. Nature. 2012 Dec 20;492(7429):382-6. doi: 10.1038/nature11737. Epub 2012 Dec 12. Nature. 2012. PMID: 23235829 Free PMC article.
References
- Ashley C T, Sutcliffe J S, Kunst C B, Leiner H A, Eichler E E, Nelson D L, Warren S T. Human and murine FMR-1: alternative splicing and translational initiation downstream of the CGG repeat. Nat Genet. 1993;4:244–251. - PubMed
- Ashley C T, Warren S T. Trinucleotide repeat expansion and human disease. Annu Rev Genet. 1995;29:703–728. - PubMed
- Ashley C T, Wilkinson K D, Reines D, Warren S T. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. - PubMed
- Borer R A, Lehner C F, Eppenberger H M, Nigg E A. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases