In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b - PubMed (original) (raw)
In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b
C L Hsieh. Mol Cell Biol. 1999 Dec.
Abstract
The putative de novo methyltransferases, Dnmt3a and Dnmt3b, were reported to have weak methyltransferase activity in methylating the 3' long terminal repeat of Moloney murine leukemia virus in vitro. The activity of these enzymes was evaluated in vivo, using a stable episomal system that employs plasmids as targets for DNA methylation in human cells. De novo methylation of a subset of the CpG sites on the stable episomes is detected in human cells overexpressing the murine Dnmt3a or Dnmt3b1 protein. This de novo methylation activity is abolished when the cysteine in the P-C motif, which is the catalytic site of cytosine methyltransferases, is replaced by a serine. The pattern of methylation on the episome is nonrandom, and different regions of the episome are methylated to different extents. Furthermore, Dnmt3a also methylates the sequence methylated by Dnmt3a on the stable episome in the corresponding chromosomal target. Overexpression of human DNMT1 or murine Dnmt3b does not lead to the same pattern or degree of de novo methylation on the episome as overexpression of murine Dnmt3a. This finding suggests that these three enzymes may have different targets or requirements, despite the fact that weak de novo methyltransferase activity has been demonstrated in vitro for all three enzymes. It is also noteworthy that both Dnmt3a and Dnmt3b proteins coat the metaphase chromosomes while displaying a more uniform pattern in the nucleus. This is the first evidence that Dnmt3a and Dnmt3b have de novo methyltransferase function in vivo and the first indication that the Dnmt3a and Dnmt3b proteins may have preferred target sites.
Figures
FIG. 1
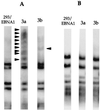
Dnmt3a and Dnmt3b expression in 293/EBNA1 cells. (A) Immunofluorescent staining of 293/EBNA1 cells transiently transfected with pMT3aMyc, pMT3bMyc, pMT3aMut, or pMT3bMut. Arrowheads indicate cells with no Myc-tagged protein expression. (B) Western blot of Myc-tagged Dnmt3a and Dnmt3b protein harvested from 3a-5, 3a-11, and 3b-11 cell clones. The solid arrowhead indicates the Dnmt3a protein, and the open arrowhead indicates the Dnmt3b protein.
FIG. 2
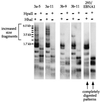
De novo methylation of assay plasmid pCLH22 by Dnmt3a and Dnmt3b. Shown is a Southern blot of pCLH22 DNA harvested 10 days after transfection from 293/EBNA, 3a, and 3b cells. The DNA was digested with restriction enzyme _Hha_I or _Hpa_II, as indicated above each lane. The probe used is the entire plasmid pCLH22. 3a-5 and 3a-11 are cell clones with stably integrated pMT3aMyc, and 3b-9 and 3b-11 are cell clones that harbor stably integrated pMT3bMyc. Plasmid DNA harvested from 293/EBNA1 cells shows the complete digestion pattern, and plasmid DNA harvested from the 3a and 3b cell clones show some fragments of increased size.
FIG. 3
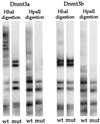
Catalytic site mutants do not methylate the assay plasmid. Shown is a Southern blot of _Hha_I- or _Hpa_II-digested plasmid DNA harvested 8 days after transfection. Plasmid expressing wild-type (wt) Dnmt3a, mutant (mut) Dnmt3a, wild-type Dnmt3b, or mutant Dnmt3b was cotransfected with assay plasmid p220.2. The p220.2 DNA harvested showed a complete _Hha_I and _Hpa_II digestion pattern when cotransfected with either pMT3aMut or pMT3bMut.
FIG. 4
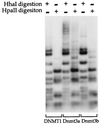
DNMT1, Dnmt3a, and Dnmt3b have distinct de novo methyltransferase activities in vivo. An expression vector for human DNMT1, murine Dnmt3a, or Dnmt3b was cotransfected into 293/EBNA1 cells with assay plasmid pCLH22. pCLH22 DNA was harvested 12 days after transfection and digested with restriction enzyme _Hha_I or _Hpa_II, as indicated above each lane. The increased-size _Hha_I and _Hpa_II fragments detected in pCLH22 DNA cotransfected with either the Dnmt3a or the Dnmt3b expression vector are the same as observed in Fig. 2. A complete digestion pattern is observed in pCLH22 DNA cotransfected with the DNMT1 expression vector, where only two very faint bands of increased size are detected in the _Hha_I- or _Hpa_II-digested DNA after long exposure.
FIG. 5
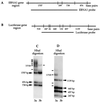
Methylation of a specific region of the episome. (A) Illustration of _Hha_I sites and _Hha_I fragment sizes within and adjacent to the EBNA1 region. (B) Illustration of _Hha_I sites and fragment sizes in the luciferase gene. (C) Southern blot of _Hha_I digestion of pCLH22 DNA harvested from transfected 3a-5 and 3b-11 cells 10 days after transfection. The ENBA1 region-specific probe used for hybridization is as indicated in panel A. All increased-size _Hha_I and _Hpa_II fragments observed in Fig. 2 are detected with this probe, as indicated with solid lines. (D) The same Southern blot in panel C, hybridized with a probe containing the luciferase coding region as indicated in panel B. The _Hha_I fragments derived from complete digestion are indicated with arrowheads labeled with fragment sizes. Four additional _Hha_I fragments were observed in the DNA harvested from 3a cells, as indicated with lines. Two of these fragments are smaller and two are larger than the largest completely digested band of 1.1 kb.
FIG. 6
Lack of methylation of plasmid without the EBNA1 sequence. (A) Southern blot of _Hha_I-digested p220.2 (lacking the luciferase gene) DNA harvested 10 days after transfection. Arrowheads indicate increased-size DNA fragments detected in 3a and 3b cells. (B) Southern blot of _Hha_I digestion of p22ΔEBNA1 DNA harvested 10 days after transfection. There is no detectable _Hha_I fragment of increased size in any cells transfected with p22ΔEBNA1. The entire plasmid is used as the probe in both panels.
FIG. 7
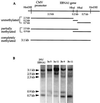
Methylation of the EBNA1 sequence in the chromosome. (A) Illustration of _Hin_dIII and _Hha_I sites within the _Hin_dIII-to-_Hin_dIII fragment from the CMV-EBNA1 construct inserted into 293 cells. DNA fragments generated from the integrated construct by a _Hin_dIII/_Hha_I double digestion of unmethylated, partially methylated, and completely methylated sequences are also illustrated. (B) Southern blot of _Hin_dIII/_Hha_I double-digested genomic DNA harvested from 293/EBNA1, 3a-5, 3a-11, 3b-9, and 3b-11 cells. The probe used is the same EBNA1 fragment as indicated in Fig. 5A. Compared with 293/EBNA1, 3b-9, and 3b-11 cells, there is a decreasing amount of DNA in the smaller fragments and increasing amount of DNA in the 3.1-kb band in the DNA extracted from 3a-5 and 3a-11 cells. Lack of fragments larger than 3.1 kb indicates complete digestion of the DNA by _Hin_dIII. Probing of the same blot by the puromycin sequence also indicates complete digestion by _Hha_I (data not shown).
Similar articles
- Murine de novo methyltransferase Dnmt3a demonstrates strand asymmetry and site preference in the methylation of DNA in vitro.
Lin IG, Han L, Taghva A, O'Brien LE, Hsieh CL. Lin IG, et al. Mol Cell Biol. 2002 Feb;22(3):704-23. doi: 10.1128/MCB.22.3.704-723.2002. Mol Cell Biol. 2002. PMID: 11784849 Free PMC article. - De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation.
Dodge JE, Ramsahoye BH, Wo ZG, Okano M, Li E. Dodge JE, et al. Gene. 2002 May 1;289(1-2):41-8. doi: 10.1016/s0378-1119(02)00469-9. Gene. 2002. PMID: 12036582 - Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b.
Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Chen T, et al. Mol Cell Biol. 2003 Aug;23(16):5594-605. doi: 10.1128/MCB.23.16.5594-5605.2003. Mol Cell Biol. 2003. PMID: 12897133 Free PMC article. - Using human disease mutations to understand de novo DNA methyltransferase function.
Rolls W, Wilson MD, Sproul D. Rolls W, et al. Biochem Soc Trans. 2024 Oct 30;52(5):2059-2075. doi: 10.1042/BST20231017. Biochem Soc Trans. 2024. PMID: 39446312 Free PMC article. Review. - Methyltransferase DNMT3B in leukemia.
Zhang H, Ying H, Wang X. Zhang H, et al. Leuk Lymphoma. 2020 Feb;61(2):263-273. doi: 10.1080/10428194.2019.1666377. Epub 2019 Sep 24. Leuk Lymphoma. 2020. PMID: 31547729 Review.
Cited by
- Systematic epigenome editing captures the context-dependent instructive function of chromatin modifications.
Policarpi C, Munafò M, Tsagkris S, Carlini V, Hackett JA. Policarpi C, et al. Nat Genet. 2024 Jun;56(6):1168-1180. doi: 10.1038/s41588-024-01706-w. Epub 2024 May 9. Nat Genet. 2024. PMID: 38724747 Free PMC article. - Exploring epigenetic strategies for the treatment of osteoporosis.
Yi SJ, Lim J, Kim K. Yi SJ, et al. Mol Biol Rep. 2024 Mar 8;51(1):398. doi: 10.1007/s11033-024-09353-4. Mol Biol Rep. 2024. PMID: 38453825 Review. - DNMT3B PWWP mutations cause hypermethylation of heterochromatin.
Taglini F, Kafetzopoulos I, Rolls W, Musialik KI, Lee HY, Zhang Y, Marenda M, Kerr L, Finan H, Rubio-Ramon C, Gautier P, Wapenaar H, Kumar D, Davidson-Smith H, Wills J, Murphy LC, Wheeler A, Wilson MD, Sproul D. Taglini F, et al. EMBO Rep. 2024 Mar;25(3):1130-1155. doi: 10.1038/s44319-024-00061-5. Epub 2024 Jan 30. EMBO Rep. 2024. PMID: 38291337 Free PMC article. - A Porcine DNMT1 Variant: Molecular Cloning and Generation of Specific Polyclonal Antibody.
Zhu L, Wang J, Zhang Y, Xiang X, Liu K, Wei J, Li Z, Shao D, Li B, Ma Z, Qiu Y. Zhu L, et al. Genes (Basel). 2023 Jun 23;14(7):1324. doi: 10.3390/genes14071324. Genes (Basel). 2023. PMID: 37510229 Free PMC article. - Epigenetic Modification of Cytosines in Hematopoietic Differentiation and Malignant Transformation.
An J, Ko M. An J, et al. Int J Mol Sci. 2023 Jan 15;24(2):1727. doi: 10.3390/ijms24021727. Int J Mol Sci. 2023. PMID: 36675240 Free PMC article. Review.
References
- Bird A P. The essentials of DNA methylation. Cell. 1992;70:5–8. - PubMed
- Cedar H. DNA methylation and gene activity. Cell. 1988;53:3–4. - PubMed
- Grawunder U, Finnie N, Jackson S P, Riwar B, Jessberger R. Expression of DNA-dependent protein kinase holoenzyme upon induction of lymphocyte differentiation and V(D)J recombination. Eur J Biochem. 1996;241:931–940. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases