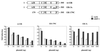Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors alpha and beta - PubMed (original) (raw)
Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors alpha and beta
C y Chang et al. Mol Cell Biol. 1999 Dec.
Abstract
Recruitment of transcriptional coactivators following ligand activation is a critical step in nuclear receptor-mediated target gene expression. Upon binding an agonist, the receptor undergoes a conformational change which facilitates the formation of a specific coactivator binding pocket within the carboxyl terminus of the receptor. This permits the alpha-helical LXXLL motif within some coactivators to interact with the nuclear receptors. Until recently, the LXXLL motif was thought to function solely as a docking module; however, it now appears that sequences flanking the core motif may play a role in determining receptor selectivity. To address this issue, we used a combinatorial phage display approach to evaluate the role of flanking sequences in influencing these interactions. We sampled more than 10(8) variations of the core LXXLL motif with estradiol-activated estrogen receptor alpha (ERalpha) as a target and found three different classes of peptides. All of these peptides interacted with ERalpha in an agonist-dependent manner and disrupted ERalpha-mediated transcriptional activity when introduced into target cells. Using a series of ERalpha-mutants, we found that these three classes of peptides showed different interaction patterns from each other, suggesting that not all LXXLL motifs are the same and that receptor binding selectivity can be achieved by altering sequences flanking the LXXLL core motif. Most notable in this regard was the discovery of a peptide which, when overexpressed in cells, selectively disrupted ERbeta- but not ERalpha-mediated reporter gene expression. This novel ERbeta-specific antagonist may be useful in identifying and characterizing the ERbeta-regulated process in estradiol-responsive cells. In conclusion, using a combinatorial approach to define cofactor-receptor interactions, we have clearly been able to demonstrate that not all LXXLL motifs are functionally equivalent, a finding which suggests that it may be possible to target receptor-LXXLL interactions to develop receptor-specific antagonists.
Figures
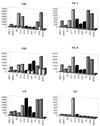
FIG. 1
Affinity selection of ERα binding motifs by using phage display technology. Baculovirus-expressed full-length ERα was treated with 10−6 M 17β-estradiol and immobilized on 96-well Immulon 4 plates as a screening target. The LXXLL motif-containing phage peptide library was constructed as described in Materials and Methods. Phage that interacted specifically with estradiol-activated ER were selected, and the peptide sequences were deduced by DNA sequencing. These peptides were classified into three different classes based on sequences flanking the conserved LXXLL motif. Peptide #293 was obtained in a similar manner from random peptide libraries; it bound specifically to estradiol-activated ERβ when analyzed in vitro. Sequences from the center three copies of LXXLL motifs in the SRC-1 and GRIP-1 coactivators are also included for comparison. For reference, we have defined the first conserved leucine as position 1.
FIG. 2
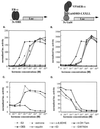
The interaction between LXXLL-containing peptides and ER occurs only in the presence of receptor agonists. The LXXLL-containing ER4 peptide sequence was fused to Gal4DBD, while the full-length ERα was expressed as a VP16 transactivation domain fusion protein. The interaction between ER4 peptide and ERα was assessed by using the 5×Gal4Luc3 reporter gene (B and D). The ability of different ER ligands to facilitate LXXLL peptide-ERα interactions was compared to the ability of these ligands to induce ER-mediated transactivation, as assayed by using the 3×ERE-TATA-Luc reporter (A and C). HepG2 cells were transiently transfected with the ERα expression vector (pRST7ERα) and its reporter 3×ERE-TATA-Luc construct (A and C) or Gal4DBD-ER4, pVP16-ERα, and 5×Gal4Luc3 (B and D) and treated with different ER ligands as indicated in the key. Luciferase (Luc) activity was normalized to the activity of the cotransfected pCMVβgal plasmid. E2, 17β-estradiol; 4-OH Tam, 4-hydroxytamoxifen; ICI, ICI 182,780; DES, diethylstilbesterol; Δ-8,9DHE, delta-8,9-dehydroestrone.
FIG. 3
Not all LXXLL peptide-ER interactions require a functional AF-2. The three groups of LXXLL-containing peptides interacted differentially with ER helix 12 mutants. (A) A schematic drawing of the wild-type (wt) ER is shown along with a region of the HBD corresponding to ER activation function 2 (AF-2). Residues that were mutated are indicated by circles. (B) Mammalian two-hybrid assays were used to test whether all the LXXLL motifs interacted with the same region of ER. Peptide sequences representing three LXXLL classes were expressed as fusion proteins to the Gal4DBD. Wild-type (wt) and mutant ERα were expressed as VP16 fusion proteins. The binding capacity of different peptides to wild-type and mutant ER was measured by using a 5×Gal4Luc3 reporter construct. GRIP-1 (NR-box) and SRC-1 (NR-box) constructs contain the center three copies of an LXXLL motif (amino acids 629 to 760 for GRIP-1 and 621 to 765 for SRC-1) fused to Gal4DBD. (C) Western analysis of the expression levels of selected Gal4DBD-peptide fusions. Nuclear extracts were prepared from transfected HeLa cells and analyzed using SDS-PAGE. The peptide-Gal4DBD fusion proteins were detected with a monoclonal antibody raised against Gal4DBD (αGal4DBD). The expression levels of the Gal4DBD fusions were normalized by assaying the levels of EGFP expressed from a cotransfected plasmid (pEGFP-C3). Specifically, the identical blot was reprobed with a polyclonal anti-GFP antibody (αGFP).
FIG. 3
Not all LXXLL peptide-ER interactions require a functional AF-2. The three groups of LXXLL-containing peptides interacted differentially with ER helix 12 mutants. (A) A schematic drawing of the wild-type (wt) ER is shown along with a region of the HBD corresponding to ER activation function 2 (AF-2). Residues that were mutated are indicated by circles. (B) Mammalian two-hybrid assays were used to test whether all the LXXLL motifs interacted with the same region of ER. Peptide sequences representing three LXXLL classes were expressed as fusion proteins to the Gal4DBD. Wild-type (wt) and mutant ERα were expressed as VP16 fusion proteins. The binding capacity of different peptides to wild-type and mutant ER was measured by using a 5×Gal4Luc3 reporter construct. GRIP-1 (NR-box) and SRC-1 (NR-box) constructs contain the center three copies of an LXXLL motif (amino acids 629 to 760 for GRIP-1 and 621 to 765 for SRC-1) fused to Gal4DBD. (C) Western analysis of the expression levels of selected Gal4DBD-peptide fusions. Nuclear extracts were prepared from transfected HeLa cells and analyzed using SDS-PAGE. The peptide-Gal4DBD fusion proteins were detected with a monoclonal antibody raised against Gal4DBD (αGal4DBD). The expression levels of the Gal4DBD fusions were normalized by assaying the levels of EGFP expressed from a cotransfected plasmid (pEGFP-C3). Specifically, the identical blot was reprobed with a polyclonal anti-GFP antibody (αGFP).
FIG. 3
Not all LXXLL peptide-ER interactions require a functional AF-2. The three groups of LXXLL-containing peptides interacted differentially with ER helix 12 mutants. (A) A schematic drawing of the wild-type (wt) ER is shown along with a region of the HBD corresponding to ER activation function 2 (AF-2). Residues that were mutated are indicated by circles. (B) Mammalian two-hybrid assays were used to test whether all the LXXLL motifs interacted with the same region of ER. Peptide sequences representing three LXXLL classes were expressed as fusion proteins to the Gal4DBD. Wild-type (wt) and mutant ERα were expressed as VP16 fusion proteins. The binding capacity of different peptides to wild-type and mutant ER was measured by using a 5×Gal4Luc3 reporter construct. GRIP-1 (NR-box) and SRC-1 (NR-box) constructs contain the center three copies of an LXXLL motif (amino acids 629 to 760 for GRIP-1 and 621 to 765 for SRC-1) fused to Gal4DBD. (C) Western analysis of the expression levels of selected Gal4DBD-peptide fusions. Nuclear extracts were prepared from transfected HeLa cells and analyzed using SDS-PAGE. The peptide-Gal4DBD fusion proteins were detected with a monoclonal antibody raised against Gal4DBD (αGal4DBD). The expression levels of the Gal4DBD fusions were normalized by assaying the levels of EGFP expressed from a cotransfected plasmid (pEGFP-C3). Specifically, the identical blot was reprobed with a polyclonal anti-GFP antibody (αGFP).
FIG. 4
The interaction of ERα with each of the three classes of LXXLL peptides identified is affected differentially by helix 12 mutations. The contributions of each of the three charged residues (D538, E542, D545) within helix 12 to LXXLL motif-ERα interactions were evaluated. Specifically, we created single point mutations of each residue to their corresponding amides and evaluated the impact of these mutations on ERα-LXXLL peptide interactions in a mammalian two-hybrid assay. The mutants indicated were generated by site-directed mutagenesis within the wild-type (wt) VP16-ERα backbone. Selected peptide sequences representing each of the three LXXLL classes were expressed as Gal4DBD fusions. The binding capacity of the different peptides to wild-type and mutant ER was measured by using a 5×Gal4Luc3 reporter construct. GRIP-1 (NR-box) and SRC-1 (NR-box) constructs contain the center three copies of an LXXLL motif (amino acids 629 to 760 for GRIP-1 and 621 to 765 for SRC-1) fused to Gal4DBD.
FIG. 5
LXXLL-containing peptides disrupt ERα transcriptional activity when overexpressed in target cells. HeLa cells were transfected with the ERα expression plasmid (pRST7ERα), 3×ERE-TATA-Luc reporter, along with increasing amounts of a construct expressing the peptide-Gal4DBD fusions as indicated. F6 contains a single copy of the F6 peptide, 2×F6 contains two copies of the F6 peptide with 50 amino acids separating the two LXXLL motifs, and GRIP-1 contains the center three NR boxes from the coactivator GRIP-1. All these peptides were expressed as fusion proteins to Gal4DBD. In addition, a pCMVβgal plasmid was cotransfected to normalize for transfection efficiency. After transfection, cells were induced with 10−7 M 17β-estradiol for 16 h before assaying. Fold induction represents the ratio of estradiol-induced activity versus no-hormone control for each transfection.
FIG. 6
The differential ability of LXXLL-containing peptides to disrupt ERα-mediated transactivation function reveals the presence of multiple ER-interacting coactivators. HepG2 cells were transfected with pRST7-ERα (wt), ERα179C, or ERα-3× mutant expression plasmids along with the 3×ERE-TATA-Luc reporter gene and increasing amounts of the Gal4DBD-peptide fusion constructs (as indicated). Fold induction represents the ratio of estradiol-induced (10−7 M) activity versus no-hormone control for each transfection.
FIG. 7
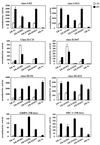
Nuclear receptors have distinct preferences for different LXXLL motifs. The interactions between different LXXLL motifs and nuclear receptors were assayed by using a mammalian two-hybrid system. Full-length receptors and selected peptides were expressed as VP16 and Gal4DBD fusion proteins, respectively. The magnitude of these interactions was measured by using a 5×Gal4Luc3 reporter gene. Open bars, no hormone; hatched or filled bars, hormone treatments. The following hormones were used in this experiment: 10−7 M 17β-estradiol for ERα and ERβ, 10−7 M progesterone for PR-A and PR-B, 10−7 M dexamethasone for GR, 10−7 M 9-_cis_-retinoic acid for RARα and RXRα, 10−7 M T3 for TRβ, 10−7 M 1,25-dihydroxyvitamin D3 for VDR, and 10−6 M 5α-dihydrotestosterone for AR. The luciferase activity was normalized to the activity of the cotransfected pCMVβgal.
FIG. 7
Nuclear receptors have distinct preferences for different LXXLL motifs. The interactions between different LXXLL motifs and nuclear receptors were assayed by using a mammalian two-hybrid system. Full-length receptors and selected peptides were expressed as VP16 and Gal4DBD fusion proteins, respectively. The magnitude of these interactions was measured by using a 5×Gal4Luc3 reporter gene. Open bars, no hormone; hatched or filled bars, hormone treatments. The following hormones were used in this experiment: 10−7 M 17β-estradiol for ERα and ERβ, 10−7 M progesterone for PR-A and PR-B, 10−7 M dexamethasone for GR, 10−7 M 9-_cis_-retinoic acid for RARα and RXRα, 10−7 M T3 for TRβ, 10−7 M 1,25-dihydroxyvitamin D3 for VDR, and 10−6 M 5α-dihydrotestosterone for AR. The luciferase activity was normalized to the activity of the cotransfected pCMVβgal.
FIG. 8
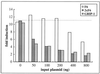
LXXLL motif-containing cofactors interact with both ERα and ERβ in vitro in a ligand-dependent manner. Equal amounts of full-length ERα, ERβ, or control BSA were immobilized on 96-well plates in the presence or absence of 1 μM estradiol. Full-length RIP140, GRIP-1, and TRAP220 were translated in vitro and labeled with [35S]methionine. Labeled cofactors were added to the wells containing immobilized protein and incubated at 4°C overnight. Unbound protein was removed by washing, and the bound protein was eluted, separated by SDS-PAGE, and visualized by autoradiography.
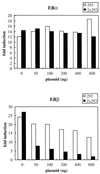
FIG. 9
Peptide #293 selectively disrupts ERβ-dependent reporter gene expression without affecting ERα-mediated transcription when expressed in target cells. Peptide #293 containing an LXXLL motif was affinity selected by phage display with estradiol-activated ERβ as a target. Expression of either one copy or two copies of this peptide did not interfere with the transcriptional activity of ERα but disrupted ERβ-mediated transcriptional activity. HeLa cells were transfected with either ERα or ERβ expression plasmids, along with 3×ERE-TATA-Luc reporter, pCMVβgal, and increasing amounts of Gal4DBD-peptide fusion constructs as indicated. Fold induction represents the ratio of activity estradiol-induced activity versus no-hormone control for each transfection.
Similar articles
- Analysis of ligand-dependent recruitment of coactivator peptides to estrogen receptor using fluorescence polarization.
Ozers MS, Ervin KM, Steffen CL, Fronczak JA, Lebakken CS, Carnahan KA, Lowery RG, Burke TJ. Ozers MS, et al. Mol Endocrinol. 2005 Jan;19(1):25-34. doi: 10.1210/me.2004-0256. Epub 2004 Sep 16. Mol Endocrinol. 2005. PMID: 15375189 - Development of peptide antagonists that target estrogen receptor beta-coactivator interactions.
Hall JM, Chang CY, McDonnell DP. Hall JM, et al. Mol Endocrinol. 2000 Dec;14(12):2010-23. doi: 10.1210/mend.14.12.0561. Mol Endocrinol. 2000. PMID: 11117531 - Selection of estrogen receptor beta- and thyroid hormone receptor beta-specific coactivator-mimetic peptides using recombinant peptide libraries.
Northrop JP, Nguyen D, Piplani S, Olivan SE, Kwan ST, Go NF, Hart CP, Schatz PJ. Northrop JP, et al. Mol Endocrinol. 2000 May;14(5):605-22. doi: 10.1210/mend.14.5.0445. Mol Endocrinol. 2000. PMID: 10809226 - Ser-884 adjacent to the LXXLL motif of coactivator TRBP defines selectivity for ERs and TRs.
Ko L, Cardona GR, Iwasaki T, Bramlett KS, Burris TP, Chin WW. Ko L, et al. Mol Endocrinol. 2002 Jan;16(1):128-40. doi: 10.1210/mend.16.1.0755. Mol Endocrinol. 2002. PMID: 11773444 - The coactivator LXXLL nuclear receptor recognition motif.
Savkur RS, Burris TP. Savkur RS, et al. J Pept Res. 2004 Mar;63(3):207-12. doi: 10.1111/j.1399-3011.2004.00126.x. J Pept Res. 2004. PMID: 15049832 Review.
Cited by
- Serine protease PRSS23 is upregulated by estrogen receptor α and associated with proliferation of breast cancer cells.
Chan HS, Chang SJ, Wang TY, Ko HJ, Lin YC, Lin KT, Chang KM, Chuang YJ. Chan HS, et al. PLoS One. 2012;7(1):e30397. doi: 10.1371/journal.pone.0030397. Epub 2012 Jan 23. PLoS One. 2012. PMID: 22291950 Free PMC article. - The normal and malignant mammary gland: a fresh look with ER beta onboard.
Warner M, Saji S, Gustafsson JA. Warner M, et al. J Mammary Gland Biol Neoplasia. 2000 Jul;5(3):289-94. doi: 10.1023/a:1009598828267. J Mammary Gland Biol Neoplasia. 2000. PMID: 14973391 Review. - Human PXR forms a tryptophan zipper-mediated homodimer.
Noble SM, Carnahan VE, Moore LB, Luntz T, Wang H, Ittoop OR, Stimmel JB, Davis-Searles PR, Watkins RE, Wisely GB, LeCluyse E, Tripathy A, McDonnell DP, Redinbo MR. Noble SM, et al. Biochemistry. 2006 Jul 18;45(28):8579-89. doi: 10.1021/bi0602821. Biochemistry. 2006. PMID: 16834332 Free PMC article. - Novel mechanism of nuclear receptor corepressor interaction dictated by activation function 2 helix determinants.
Moraitis AN, Giguère V, Thompson CC. Moraitis AN, et al. Mol Cell Biol. 2002 Oct;22(19):6831-41. doi: 10.1128/MCB.22.19.6831-6841.2002. Mol Cell Biol. 2002. PMID: 12215540 Free PMC article. - Estrogen receptor transcription and transactivation: Estrogen receptor alpha and estrogen receptor beta: regulation by selective estrogen receptor modulators and importance in breast cancer.
Katzenellenbogen BS, Katzenellenbogen JA. Katzenellenbogen BS, et al. Breast Cancer Res. 2000;2(5):335-44. doi: 10.1186/bcr78. Epub 2000 Jul 7. Breast Cancer Res. 2000. PMID: 11250726 Free PMC article. Review.
References
- Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. - PubMed
- Berrevoets C A, Doesburg P, Steketee K, Trapman J, Brinkmann A O. Functional interactions of the AF-2 activation domain core region of the human androgen receptor with the amino-terminal domain and with the transcriptional coactivator TIF2 (transcriptional intermediary factor2) Mol Endocrinol. 1998;12:1172–1183. - PubMed
- Brzozowski A M, Pike A C, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J A, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials