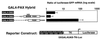Paired-homeodomain transcription factor PAX4 acts as a transcriptional repressor in early pancreatic development - PubMed (original) (raw)
Paired-homeodomain transcription factor PAX4 acts as a transcriptional repressor in early pancreatic development
S B Smith et al. Mol Cell Biol. 1999 Dec.
Abstract
The paired-homeodomain transcription factor PAX4 is expressed in the developing pancreas and along with PAX6 is required for normal development of the endocrine cells. In the absence of PAX4, the numbers of insulin-producing beta cells and somatostatin-producing delta cells are drastically reduced, while the numbers of glucagon-producing alpha cells are increased. To gain insight into PAX4 function, we cloned a full-length Pax4 cDNA from a beta-cell cDNA library and identified a bipartite consensus DNA binding sequence consisting of a homeodomain binding site separated from a paired domain binding site by 15 nucleotides. The paired half of this consensus sequence has similarities to the PAX6 paired domain consensus binding site, and the two proteins bind to common sequences in several islet genes, although with different relative affinities. When expressed in an alpha-cell line, PAX4 represses transcription through the glucagon or insulin promoters or through an isolated PAX4 binding site. This repression is not simply due to competition with the PAX6 transcriptional activator for the same binding site, since PAX4 fused to the unrelated yeast GAL4 DNA binding domain also represses transcription through the GAL4 binding site in the alpha-cell line and to a lesser degree in beta-cell lines and NIH 3T3 cells. Repressor activity maps to more than one domain within the molecule, although the homeodomain and carboxyl terminus give the strongest repression. PAX4 transcriptional regulation apparently plays a role only early in islet development, since Pax4 mRNA as determined by reverse transcriptase PCR peaks at embryonic day 13.5 in the fetal mouse pancreas and is undetectable in adult islets. In summary, PAX4 can function as a transcriptional repressor and is expressed early in pancreatic development, which may allow it to suppress alpha-cell differentiation and permit beta-cell differentiation.
Figures
FIG. 1
PAX4 binding site selection. In vitro-produced truncated PAX4 protein including the paired domain and homeodomain was used in EMSA to select sequences with high binding affinity from a random pool of oligonucleotides containing 30 degenerate base pairs flanked by PCR primer sites. (A) Alignment of sequences of oligonucleotide products subcloned and sequenced after 13 rounds of selection. Sequences are aligned by the MacVector multiple-sequence alignment function to show the best fit. (B) Alignment of homeodomain type sequences after 13 rounds of selection. (C) Alignment of paired domain-type sequences after 13 rounds of selection. (D) Alignment of oligonucleotide products subcloned and sequenced after 10 rounds of selection. TAAT sequences are underlined. (E) The consensus PAX4 recognition sequence established from the aligned sequences compared to the PAX6 consensus recognition sequence, based on paired domain binding alone (7). Matching bases are in boldface; rat hormone gene regulatory elements with possible PAX4 binding sites are underlined.
FIG. 2
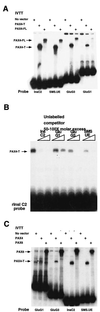
PAX4 binding to islet gene promoters. (A) EMSA using truncated (PAX4-T) and full-length (PAX4-FL) PAX4 to bind to hormone gene regulatory elements. 32P-end-labeled oligonucleotides (10,000 cpm) were incubated with 1 μl of a 50-μl IVTT reaction in each lane and then electrophoresed. (B) EMSA using PAX4-T to bind to 32P-end-labeled rInsIC2 probe (10,000 cpm). A 50- to 100-fold molar excess of unlabeled competitor oligonucleotides was added to the lanes indicated; 0.05 μl of a 50-μl IVTT reaction was used in each lane. (C) EMSA comparing the binding of PAX4-T and PAX6 to 32P-end-labeled hormone gene regulatory elements; 0.1 μl of a 50-μl IVTT reaction was used in each lane. The open arrowhead (>) points to a weak PAX4-T–GluG1 complex.
FIG. 3
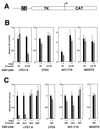
PAX4 transcriptional repression. Transient transfections of the indicated cell lines were performed with the PAX4 expression vector and reporter constructs consisting of the HSV-TK promoter alone (TK) or the isolated rInsIC2 element linked to TK-HSV (C2-TK) (A) and the −410-bp rat insulin I promoter (RIP), −482-bp rat glucagon promoter (GLU), RSV LTR (RSV), or CMV promoter (CMV) driving CAT expression. Transfections were performed in duplicate on at least three separate occasions. Relative CAT activity of the TK-CAT reporter in the absence of PAX4 expression is set arbitrarily at +1. In panel B, relative CAT activity of each construct in the absence of PAX4 expression is set arbitrarily at +1. Graphs show mean ± standard error of the mean.
FIG. 4
PAX4 repressor domains. (A) Comparison of PAX4 and PAX6 structure. PD, paired domain; HD, homeodomain. (B) Transient transfections of the indicated cell lines were performed with the chimeric PAX4-GAL4 DBD fusion protein constructs shown and a reporter construct consisting of the GAL4 binding site linked to HSV-TK driving CAT expression (GAL4UAS-TK-CAT). (C) Transient transfections of βTC3 and NIH3T3 cells were performed with a reporter construct consisting of five copies of the GAL4 GAL4 UAS. (D) Transient transfections of αTC1.6 cells were performed with a reporter construct consisting of HSV-TK driving CAT expression without a GAL4 binding site (TK-CAT). (E) Transient transfections of βTC3 cells were performed with a reporter consisting of five GAL4 binding sites linked to the minimal adenovirus E1b promoter driving CAT expression (5XGAL4UAS-E1b-CAT). Note that the lowermost expression construct consists of the PAX6 C-terminal domain fused to the GAL4 DBD. Transfections were performed in duplicate on at least three separate occasions. Relative CAT activity of the GAL4-DBD expression construct alone is set arbitrarily at +1. Graphs show mean ± standard error of the mean.
FIG. 5
Expression of Pax4 mRNA. RT-PCR was performed with 250 ng of total RNA from the tissues shown. PCR products were removed and separated by polyacrylamide gel electrophoresis after 30 cycles for PAX4 and β-actin, and 25 cycles for insulin II. Lanes labeled-RT show the product of identical samples performed without reverse transcriptase using RNA from e13.5 fetal mouse pancreas (A) and βTC3 cells (B).
FIG. 6
PAX4 transcriptional repression in fetal pancreatic epithelium. Transient transfections were performed in mouse e11.5 pancreatic epithelium with expression vectors containing the GAL4 DBD coding sequence fused with the PAX4 and PAX6 cDNA fragments shown, the pFOXLuc2.TK.5GAL reporter plasmid, and the pFOXEGFP.CMV internal standard. Transfections were performed in triplicate on individual fetal pancreatic buds. Luciferase mRNA levels were measured by RT-PCR and standardized to EGFP mRNA levels. The graph shows mean ± standard error of the mean.
Similar articles
- Pax4 represses pancreatic glucagon gene expression.
Petersen HV, Jørgensen MC, Andersen FG, Jensen J, F-Nielsen T, Jørgensen R, Madsen OD, Serup P. Petersen HV, et al. Mol Cell Biol Res Commun. 2000 Apr;3(4):249-54. doi: 10.1006/mcbr.2000.0220. Mol Cell Biol Res Commun. 2000. PMID: 10891400 - Identification of a portable repression domain and an E1A-responsive activation domain in Pax4: a possible role of Pax4 as a transcriptional repressor in the pancreas.
Fujitani Y, Kajimoto Y, Yasuda T, Matsuoka TA, Kaneto H, Umayahara Y, Fujita N, Watada H, Miyazaki JI, Yamasaki Y, Hori M. Fujitani Y, et al. Mol Cell Biol. 1999 Dec;19(12):8281-91. doi: 10.1128/MCB.19.12.8281. Mol Cell Biol. 1999. PMID: 10567553 Free PMC article. - Regulation of Pax4 paired homeodomain gene by neuron-restrictive silencer factor.
Kemp DM, Lin JC, Habener JF. Kemp DM, et al. J Biol Chem. 2003 Sep 12;278(37):35057-62. doi: 10.1074/jbc.M305891200. Epub 2003 Jun 26. J Biol Chem. 2003. PMID: 12829700 - Glucagon gene expression in the endocrine pancreas: the role of the transcription factor Pax6 in α-cell differentiation, glucagon biosynthesis and secretion.
Gosmain Y, Cheyssac C, Heddad Masson M, Dibner C, Philippe J. Gosmain Y, et al. Diabetes Obes Metab. 2011 Oct;13 Suppl 1:31-8. doi: 10.1111/j.1463-1326.2011.01445.x. Diabetes Obes Metab. 2011. PMID: 21824254 Review. - The gene Pax4 is an essential regulator of pancreatic beta-cell development.
Sosa-Pineda B. Sosa-Pineda B. Mol Cells. 2004 Dec 31;18(3):289-94. Mol Cells. 2004. PMID: 15650323 Review.
Cited by
- Homeobox genes in the rodent pineal gland: roles in development and phenotype maintenance.
Rath MF, Rohde K, Klein DC, Møller M. Rath MF, et al. Neurochem Res. 2013 Jun;38(6):1100-12. doi: 10.1007/s11064-012-0906-y. Epub 2012 Oct 18. Neurochem Res. 2013. PMID: 23076630 Free PMC article. Review. - Pax4 is not essential for beta-cell differentiation in zebrafish embryos but modulates alpha-cell generation by repressing arx gene expression.
Djiotsa J, Verbruggen V, Giacomotto J, Ishibashi M, Manning E, Rinkwitz S, Manfroid I, Voz ML, Peers B. Djiotsa J, et al. BMC Dev Biol. 2012 Dec 17;12:37. doi: 10.1186/1471-213X-12-37. BMC Dev Biol. 2012. PMID: 23244389 Free PMC article. - The activation of the rat insulin gene II by BETA2 and PDX-1 in rat insulinoma cells is repressed by Pax6.
Wolf G, Hessabi B, Karkour A, Henrion U, Dahlhaus M, Ostmann A, Giese B, Fraunholz M, Grabarczyk P, Jack R, Walther R. Wolf G, et al. Mol Endocrinol. 2010 Dec;24(12):2331-42. doi: 10.1210/me.2009-0220. Epub 2010 Oct 13. Mol Endocrinol. 2010. PMID: 20943817 Free PMC article. - Evolutionary conservation of glucose-dependent insulinotropic polypeptide (GIP) gene regulation and the enteroinsular axis.
Musson MC, Jepeal LI, Sharifnia T, Wolfe MM. Musson MC, et al. Regul Pept. 2010 Sep 24;164(2-3):97-104. doi: 10.1016/j.regpep.2010.05.007. Epub 2010 Jun 2. Regul Pept. 2010. PMID: 20621665 Free PMC article. - Stem cell therapy to treat diabetes mellitus.
Liew CG, Andrews PW. Liew CG, et al. Rev Diabet Stud. 2008 Winter;5(4):203-19. doi: 10.1900/RDS.2008.5.203. Epub 2009 Feb 10. Rev Diabet Stud. 2008. PMID: 19290381 Free PMC article.
References
- Ahlgren U, Jonsson J, Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. - PubMed
- Ahlgren U, Pfaff S L, Jessell T M, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–260. - PubMed
- Blackwell T K, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. - PubMed
- Czerny T, Schaffner G, Busslinger M. DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev. 1993;7:2048–2061. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases