Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis - PubMed (original) (raw)
Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis
M Achtman et al. Proc Natl Acad Sci U S A. 1999.
Erratum in
- Proc Natl Acad Sci U S A 2000 Jul 5;97(14):8192
Abstract
Plague, one of the most devastating diseases of human history, is caused by Yersinia pestis. In this study, we analyzed the population genetic structure of Y. pestis and the two other pathogenic Yersinia species, Y. pseudotuberculosis and Y. enterocolitica. Fragments of five housekeeping genes and a gene involved in the synthesis of lipopolysaccharide were sequenced from 36 strains representing the global diversity of Y. pestis and from 12-13 strains from each of the other species. No sequence diversity was found in any Y. pestis gene, and these alleles were identical or nearly identical to alleles from Y. pseudotuberculosis. Thus, Y. pestis is a clone that evolved from Y. pseudotuberculosis 1,500-20,000 years ago, shortly before the first known pandemics of human plague. Three biovars (Antiqua, Medievalis, and Orientalis) have been distinguished by microbiologists within the Y. pestis clone. These biovars form distinct branches of a phylogenetic tree based on restriction fragment length polymorphisms of the locations of the IS100 insertion element. These data are consistent with previous inferences that Antiqua caused a plague pandemic in the sixth century, Medievalis caused the Black Death and subsequent epidemics during the second pandemic wave, and Orientalis caused the current plague pandemic.
Figures
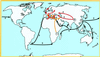
Figure 1
Routes followed by the three plague pandemic waves, labeled 1, 2, and 3. Circled numbers indicate the regions thought to be the origins of these pandemic waves.
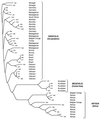
Figure 2
Neighbor-joining phylogenetic tree of band patterns of IS_100_ in 49 strains of Y. pestis.
Similar articles
- Analysis of the three Yersinia pestis CRISPR loci provides new tools for phylogenetic studies and possibly for the investigation of ancient DNA.
Vergnaud G, Li Y, Gorgé O, Cui Y, Song Y, Zhou D, Grissa I, Dentovskaya SV, Platonov ME, Rakin A, Balakhonov SV, Neubauer H, Pourcel C, Anisimov AP, Yang R. Vergnaud G, et al. Adv Exp Med Biol. 2007;603:327-38. doi: 10.1007/978-0-387-72124-8_30. Adv Exp Med Biol. 2007. PMID: 17966429 - Genome plasticity in Yersinia pestis.
Radnedge L, Agron PG, Worsham PL, Andersen GL. Radnedge L, et al. Microbiology (Reading). 2002 Jun;148(Pt 6):1687-1698. doi: 10.1099/00221287-148-6-1687. Microbiology (Reading). 2002. PMID: 12055289 - Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis.
Hinchliffe SJ, Isherwood KE, Stabler RA, Prentice MB, Rakin A, Nichols RA, Oyston PC, Hinds J, Titball RW, Wren BW. Hinchliffe SJ, et al. Genome Res. 2003 Sep;13(9):2018-29. doi: 10.1101/gr.1507303. Genome Res. 2003. PMID: 12952873 Free PMC article. - Plague in the genomic area.
Drancourt M. Drancourt M. Clin Microbiol Infect. 2012 Mar;18(3):224-30. doi: 10.1111/j.1469-0691.2012.03774.x. Clin Microbiol Infect. 2012. PMID: 22369155 Review. - Genome and Evolution of Yersinia pestis.
Cui Y, Song Y. Cui Y, et al. Adv Exp Med Biol. 2016;918:171-192. doi: 10.1007/978-94-024-0890-4_6. Adv Exp Med Biol. 2016. PMID: 27722863 Review.
Cited by
- Subtle genetic modifications transformed an enteropathogen into a flea-borne pathogen.
Carniel E. Carniel E. Proc Natl Acad Sci U S A. 2014 Dec 30;111(52):18409-10. doi: 10.1073/pnas.1421887112. Epub 2014 Dec 22. Proc Natl Acad Sci U S A. 2014. PMID: 25535337 Free PMC article. No abstract available. - Draft Genome Sequences of Two Yersinia pseudotuberculosis ST43 (O:1b) Strains, B-7194 and B-7195.
Blouin Y, Platonov ME, Pourcel C, Evseeva VV, Afanas'ev MV, Balakhonov SV, Anisimov AP, Vergnaud G. Blouin Y, et al. Genome Announc. 2013 Jul 18;1(4):e00510-13. doi: 10.1128/genomeA.00510-13. Genome Announc. 2013. PMID: 23868131 Free PMC article. - Bacterial phylogenetic tree construction based on genomic translation stop signals.
Xu L, Kuo J, Liu JK, Wong TY. Xu L, et al. Microb Inform Exp. 2012 May 31;2(1):6. doi: 10.1186/2042-5783-2-6. Microb Inform Exp. 2012. PMID: 22651236 Free PMC article. - Ancient familial Mediterranean fever mutations in human pyrin and resistance to Yersinia pestis.
Park YH, Remmers EF, Lee W, Ombrello AK, Chung LK, Shilei Z, Stone DL, Ivanov MI, Loeven NA, Barron KS, Hoffmann P, Nehrebecky M, Akkaya-Ulum YZ, Sag E, Balci-Peynircioglu B, Aksentijevich I, Gül A, Rotimi CN, Chen H, Bliska JB, Ozen S, Kastner DL, Shriner D, Chae JJ. Park YH, et al. Nat Immunol. 2020 Aug;21(8):857-867. doi: 10.1038/s41590-020-0705-6. Epub 2020 Jun 29. Nat Immunol. 2020. PMID: 32601469 Free PMC article. - The twin arginine translocation system is essential for virulence of Yersinia pseudotuberculosis.
Lavander M, Ericsson SK, Bröms JE, Forsberg A. Lavander M, et al. Infect Immun. 2006 Mar;74(3):1768-76. doi: 10.1128/IAI.74.3.1768-1776.2006. Infect Immun. 2006. PMID: 16495550 Free PMC article.
References
- Brossollet J, Mollaret H. Pourquoi la peste? Le rat, la puce et le bubon. Paris, France: Gallimard; 1994.
- Yersin A. Ann Inst Pasteur. 1894;2:428–430.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical