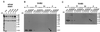Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens - PubMed (original) (raw)
Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens
H Schmidt-Eisenlohr et al. J Bacteriol. 1999 Dec.
Abstract
Three VirB proteins (VirB1*, VirB2, and VirB5) have been implicated as putative components of the T pilus from Agrobacterium tumefaciens, which likely mediates binding to plant cells followed by transfer of genetic material. Recently, VirB2 was indeed shown to be its major component (E.-M. Lai and C. I. Kado, J. Bacteriol. 180:2711-2717, 1998). Here, the influence of other Vir proteins on the stability and cellular localization of VirB1*, VirB2, and VirB5 was analyzed. Solubility of VirB1* and membrane association of VirB2 proved to be inherent features of these proteins, independent of virulence gene induction. In contrast, cellular levels of VirB5 were strongly reduced in the absence of other Vir proteins, indicating its stabilization by protein-protein interactions. The assembly and composition of the T pilus were analyzed in nopaline strain C58(pTiC58), its flagellum-free derivative NT1REB(pJK270), and octopine strain A348(pTiA6) following optimized virulence gene induction on solid agar medium. In all strains VirB2 was the major pilus component and VirB5 cofractionated during several purification steps, such as ultracentrifugation, gel filtration, and sucrose gradient centrifugation. VirB5 may therefore be directly involved in pilus assembly, possibly as minor component. In contrast, secreted VirB1* showed no association with the T pilus. In-frame deletions in genes virB1, virB2, virB5, and virB6 blocked the formation of virulence gene-dependent extracellular high-molecular-weight structures. Thus, an intact VirB machinery as well as VirB2 and VirB5 are required for T-pilus formation.
Figures
FIG. 1
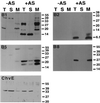
Cellular localization of VirB1, VirB2, VirB5, VirB8, and ChvE in strain C58. Western blot analysis with different specific antisera after SDS-PAGE of subcellular fractions from agrobacteria grown on AB minimal medium plates under virulence gene-inducing (+AS) and noninducing (−AS) conditions is shown. Lanes: T, total cell lysate; S, soluble fraction; M, membrane proteins. ChvE is not encoded by the virulence regulon and therefore is detected in lysates from cells grown under both conditions. Numbers on the right are molecular weights in thousands.
FIG. 2
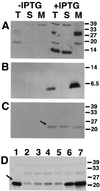
Cellular localization of VirB1, VirB2, and VirB5 in the absence of Vir proteins. Western blot analysis with specific antisera after SDS-PAGE of cell lysates from strains CB1001(pTrcB1) (A), PC1002(pTrcB2) (B), and CB1005(pTrcB5) (C) grown on AB minimal medium plates in the presence of IPTG (0.5 mM) for induction is shown. Lanes: T, total cell lysate; S, soluble fraction; M, membrane proteins. (D) Cellular levels of VirB5 in strain C58 (lane 1) (with AS) and in strain CB1005(pTrcB5) grown in the presence of IPTG and the following concentrations (micromolar) of AS for virulence gene induction: 0 (lane 2), 0.01 (lane 3), 0.1 (lane 4), 1 (lane 5), 10 (lane 6), and 200 (lane 7). Detection of VirB5 is indicated by arrows. Numbers on the right are molecular weights in thousands.
FIG. 3
Isolation of T pili from the surface of nopaline strain C58. High-molecular-weight structures were isolated by shearing from the surface of agrobacteria grown on AB minimal medium plates under virulence gene-inducing (+AS) and noninducing (−AS) conditions. The resulting samples C (cells), S1 (supernatant after shearing), S2 (supernatant after high-speed centrifugation), and P (pellet after high-speed centrifugation) were subjected to SDS-PAGE and analyzed by silver staining (A) or Western blotting with specific antisera for VirB1 (B), VirB2 (C), or VirB5 (D). VirB components detected in the pellet after ultracentrifugation are indicated by arrows. Numbers on the right are molecular weights in thousands.
FIG. 4
Ultrastructural analysis of extracellular high-molecular-weight structures. Transmission electron microscopic analysis of T pili and flagella isolated from strain C58 is shown. (A) Comparison of flagellum (asterisk) and T pilus (arrow); (B) low-magnification image of long T-pilus fragment; (C) bundle of T pili with terminal sacculi (arrowhead); (D) higher magnification of sacculus-like structure.
FIG. 5
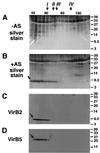
Isolation of the T pili from the surface of octopine strain A348 and virB deletion mutants PC1001 (virB1 deletion), PC1002 (virB2 deletion), PC1005 (virB5 deletion), and PC1006 (virB6 deletion). High-molecular-weight structures were isolated by shearing from the surface of agrobacteria grown on AB minimal medium plates under virulence gene-inducing (+AS) and noninducing (−AS) conditions. The resulting samples C (cells), S1 (supernatant after shearing), S2 (supernatant after high speed centrifugation), and P (pellet after high-speed centrifugation) were subjected to SDS-PAGE. Pellet fractions were analyzed by silver staining (A). Western blotting with specific VirB2 (B) or VirB5 (C) antisera was used to monitor T-pilus purification. VirB components detected in the pellet after ultracentrifugation are indicated by arrows. Numbers on the right are molecular weights in thousands.
FIG. 6
VirB2 and VirB5 elute in a high-molecular-weight structure from a Superdex 200 gel filtration column. Surface structures isolated by high-speed centrifugation from agrobacteria grown under virulence gene-inducing (+AS) and noninducing (−AS) conditions were subjected to gel filtration chromatography. Column fractions were subjected to SDS-PAGE followed by silver staining (A and B) or Western blotting with specific antisera for VirB2 (C) or VirB5 (D). VirB2 and VirB5 are indicated by arrows. Molecular weights and elution volumes of reference proteins for calibration of the gel filtration column: I, ferritin (440,000 and 63 ml); II, aldolase (158,000 and 70.4 ml); III, bovine serum albumin (68,000 and 76.8 ml); IV, cytochrome c (12,000 and 92.8 ml). Numbers on the right are molecular weights in thousands.
Similar articles
- VirB1* promotes T-pilus formation in the vir-Type IV secretion system of Agrobacterium tumefaciens.
Zupan J, Hackworth CA, Aguilar J, Ward D, Zambryski P. Zupan J, et al. J Bacteriol. 2007 Sep;189(18):6551-63. doi: 10.1128/JB.00480-07. Epub 2007 Jul 13. J Bacteriol. 2007. PMID: 17631630 Free PMC article. - VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus.
Sagulenko V, Sagulenko E, Jakubowski S, Spudich E, Christie PJ. Sagulenko V, et al. J Bacteriol. 2001 Jun;183(12):3642-51. doi: 10.1128/JB.183.12.3642-3651.2001. J Bacteriol. 2001. PMID: 11371529 Free PMC article. - Assembly of the VirB transport complex for DNA transfer from Agrobacterium tumefaciens to plant cells.
Zupan JR, Ward D, Zambryski P. Zupan JR, et al. Curr Opin Microbiol. 1998 Dec;1(6):649-55. doi: 10.1016/s1369-5274(98)80110-0. Curr Opin Microbiol. 1998. PMID: 10066547 Review. - The role of the T-pilus in horizontal gene transfer and tumorigenesis.
Kado CI. Kado CI. Curr Opin Microbiol. 2000 Dec;3(6):643-8. doi: 10.1016/s1369-5274(00)00154-5. Curr Opin Microbiol. 2000. PMID: 11121787 Review.
Cited by
- Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori.
Terradot L, Bayliss R, Oomen C, Leonard GA, Baron C, Waksman G. Terradot L, et al. Proc Natl Acad Sci U S A. 2005 Mar 22;102(12):4596-601. doi: 10.1073/pnas.0408927102. Epub 2005 Mar 11. Proc Natl Acad Sci U S A. 2005. PMID: 15764702 Free PMC article. - The N- and C-terminal portions of the Agrobacterium VirB1 protein independently enhance tumorigenesis.
Llosa M, Zupan J, Baron C, Zambryski P. Llosa M, et al. J Bacteriol. 2000 Jun;182(12):3437-45. doi: 10.1128/JB.182.12.3437-3445.2000. J Bacteriol. 2000. PMID: 10852875 Free PMC article. - Agrobacterium tumefaciens Type IV and Type VI Secretion Systems Reside in Detergent-Resistant Membranes.
Czolkoss S, Safronov X, Rexroth S, Knoke LR, Aktas M, Narberhaus F. Czolkoss S, et al. Front Microbiol. 2021 Nov 25;12:754486. doi: 10.3389/fmicb.2021.754486. eCollection 2021. Front Microbiol. 2021. PMID: 34899640 Free PMC article. - Recent advances in the structural and molecular biology of type IV secretion systems.
Trokter M, Felisberto-Rodrigues C, Christie PJ, Waksman G. Trokter M, et al. Curr Opin Struct Biol. 2014 Aug;27:16-23. doi: 10.1016/j.sbi.2014.02.006. Epub 2014 Apr 5. Curr Opin Struct Biol. 2014. PMID: 24709394 Free PMC article. Review. - Expression and physiological relevance of Agrobacterium tumefaciens phosphatidylcholine biosynthesis genes.
Klüsener S, Aktas M, Thormann KM, Wessel M, Narberhaus F. Klüsener S, et al. J Bacteriol. 2009 Jan;191(1):365-74. doi: 10.1128/JB.01183-08. Epub 2008 Oct 31. J Bacteriol. 2009. PMID: 18978052 Free PMC article.
References
- Anthony K G, Sherbourne C, Sherburne R, Frost L S. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol Microbiol. 1994;13:939–953. - PubMed
- Baron C, Zambryski P C. Plant transformation: a pilus in Agrobacterium T-DNA transfer. Curr Biol. 1996;6:1567–1569. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources