Measurement of locus copy number by hybridisation with amplifiable probes - PubMed (original) (raw)
Measurement of locus copy number by hybridisation with amplifiable probes
J A Armour et al. Nucleic Acids Res. 2000.
Abstract
Despite its fundamental importance in genome analysis, it is only recently that systematic approaches have been developed to assess copy number at specific genetic loci, or to examine genomic DNA for submicro-scopic deletions of unknown location. In this report we show that short probes can be recovered and amplified quantitatively following hybridisation to genomic DNA. This simple observation forms the basis of a new approach to determining locus copy number in complex genomes. The power and specificity of multiplex amplifiable probe hybridisation is demonstrated by the simultaneous assessment of copy number at a set of 40 human loci, including detection of deletions causing Duchenne muscular dystrophy and Prader-Willi/Angelman syndromes. Assembly of other probe sets will allow novel, technically simple approaches to a wide variety of genetic analyses, including the potential for extension to high resolution genome-wide screens for deletions and amplifications.
Figures
Figure 1
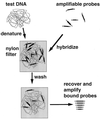
General principle of MAPH. Test DNA is denatured and hybridised with a set of amplifiable probes, each recognising a unique region in the genome. After stringent washing to allow retention only of specifically bound probes, the bound probe fragments are amplified using a common primer pair and quantified, for example after polyacrylamide gel electrophoresis. Since an excess of probe is used, the amount of each probe recovered will be proportional to the copy number of the corresponding sequence in the test DNA.
Figure 2
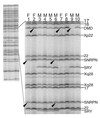
Gel analysis of recovered probes from analysis of DNA from 10 individuals examined using a mix of 40 probes from human loci, of which 13 are shown in the enlarged region. The chromosome or locus of origin for each probe is shown on the right. Arrows mark deleted bands (see also Fig. 3): individual 9 is a male with a complete deletion of exon 53 of DMD, and heterozygous deletions of this region of DMD can be seen in females 2 and 7 as reductions in the intensity relative to the adjacent Xp22 probe. Individuals 1 and 6 have heterozygous deletions of SNRPN.
Figure 3
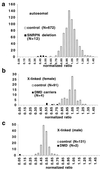
Quantitative analysis of signal intensity from two separate experiments each examining 12 DNA samples (the 10 individuals shown in Fig. 2 plus two further control samples) with a total of 40 probes. Phosphorimager data were used to produce estimates of relative intensity for each band, with the mean for each probe standardised to 1.0 (see Materials and Methods). Among control samples, ratios centre on 1.0 for autosomal probes and X-linked probes in females, and are clustered around 0.5 for X-linked probes in males.
Similar articles
- High throughput screening of human subtelomeric DNA for copy number changes using multiplex amplifiable probe hybridisation (MAPH).
Hollox EJ, Atia T, Cross G, Parkin T, Armour JA. Hollox EJ, et al. J Med Genet. 2002 Nov;39(11):790-5. doi: 10.1136/jmg.39.11.790. J Med Genet. 2002. PMID: 12414816 Free PMC article. - High-resolution analysis of acquired genomic imbalances in bone marrow samples from chronic myeloid leukemia patients by use of multiple short DNA probes.
Reid AG, Tarpey PS, Nacheva EP. Reid AG, et al. Genes Chromosomes Cancer. 2003 Jul;37(3):282-90. doi: 10.1002/gcc.10215. Genes Chromosomes Cancer. 2003. PMID: 12759926 - High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays.
Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, Gray JW, Albertson DG. Pinkel D, et al. Nat Genet. 1998 Oct;20(2):207-11. doi: 10.1038/2524. Nat Genet. 1998. PMID: 9771718 - Multiplex Amplifiable Probe Hybridization (MAPH) methodology as an alternative to comparative genomic hybridization (CGH).
Kousoulidou L, Sismani C, Patsalis PC. Kousoulidou L, et al. Methods Mol Biol. 2010;653:47-71. doi: 10.1007/978-1-60761-759-4_4. Methods Mol Biol. 2010. PMID: 20721737 Review. - MLPA and MAPH: sensitive detection of deletions and duplications.
den Dunnen JT, White SJ. den Dunnen JT, et al. Curr Protoc Hum Genet. 2006 Nov;Chapter 7:Unit 7.14. doi: 10.1002/0471142905.hg0714s51. Curr Protoc Hum Genet. 2006. PMID: 18428396 Review.
Cited by
- Copy number variation analysis by ligation-dependent PCR based on magnetic nanoparticles and chemiluminescence.
Liu M, Hu P, Zhang G, Zeng Y, Yang H, Fan J, Jin L, Liu H, Deng Y, Li S, Zeng X, Elingarami S, He N. Liu M, et al. Theranostics. 2015 Jan 1;5(1):71-85. doi: 10.7150/thno.10117. eCollection 2015. Theranostics. 2015. PMID: 25553099 Free PMC article. - Clinical impact of copy number variation analysis using high-resolution microarray technologies: advantages, limitations and concerns.
Coughlin CR 2nd, Scharer GH, Shaikh TH. Coughlin CR 2nd, et al. Genome Med. 2012 Oct 30;4(10):80. doi: 10.1186/gm381. eCollection 2012. Genome Med. 2012. PMID: 23114084 Free PMC article. Review. - Exon array CGH: detection of copy-number changes at the resolution of individual exons in the human genome.
Dhami P, Coffey AJ, Abbs S, Vermeesch JR, Dumanski JP, Woodward KJ, Andrews RM, Langford C, Vetrie D. Dhami P, et al. Am J Hum Genet. 2005 May;76(5):750-62. doi: 10.1086/429588. Epub 2005 Mar 8. Am J Hum Genet. 2005. PMID: 15756638 Free PMC article. - Comprehensive detection of genomic duplications and deletions in the DMD gene, by use of multiplex amplifiable probe hybridization.
White S, Kalf M, Liu Q, Villerius M, Engelsma D, Kriek M, Vollebregt E, Bakker B, van Ommen GJ, Breuning MH, den Dunnen JT. White S, et al. Am J Hum Genet. 2002 Aug;71(2):365-74. doi: 10.1086/341942. Epub 2002 Jul 8. Am J Hum Genet. 2002. PMID: 12111668 Free PMC article. - Copy number variation and disease resistance in plants.
Dolatabadian A, Patel DA, Edwards D, Batley J. Dolatabadian A, et al. Theor Appl Genet. 2017 Dec;130(12):2479-2490. doi: 10.1007/s00122-017-2993-2. Epub 2017 Oct 17. Theor Appl Genet. 2017. PMID: 29043379 Review.
References
- Baker S.J., Fearon,E.R., Nigro,J.M., Hamilton,S.R., Preisinger,A.C., Jessup,J.M., van Tuinen,P., Ledbetter,D.H., Barker,D.F., Nakamura,Y., White,R. and Vogelstein,B. (1989) Science, 244, 217–222. - PubMed
- Fearon E., Cho,K.R., Nigro,J.M., Kern,S.E., Simons,J.W., Ruppert,J.M., Hamilton,S.R., Preisinger,A.C., Thomas,G., Kinzler,K.W. and Vogelstein,B. (1990) Science, 247, 49–56. - PubMed
- Vogelstein B., Fearon,E.R., Kern,S.E., Hamilton,S.R., Preisinger,A.C., Nakamura,Y. and White,R. (1989) Science, 244, 207–211. - PubMed
- Solomon E., Borrow,J. and Goddard,A.D. (1991) Science, 254, 1153–1160. - PubMed
- Flint J., Wilkie,A.O.M., Buckle,V.J., Winter,R.M., Holland,A.J. and McDermid,H.E. (1995) Nature Genet., 9, 132–139. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources