A phylogenomic study of DNA repair genes, proteins, and processes - PubMed (original) (raw)
Comparative Study
A phylogenomic study of DNA repair genes, proteins, and processes
J A Eisen et al. Mutat Res. 1999.
Abstract
The ability to recognize and repair abnormal DNA structures is common to all forms of life. Studies in a variety of species have identified an incredible diversity of DNA repair pathways. Documenting and characterizing the similarities and differences in repair between species has important value for understanding the origin and evolution of repair pathways as well as for improving our understanding of phenotypes affected by repair (e.g., mutation rates, lifespan, tumorigenesis, survival in extreme environments). Unfortunately, while repair processes have been studied in quite a few species, the ecological and evolutionary diversity of such studies has been limited. Complete genome sequences can provide potential sources of new information about repair in different species. In this paper, we present a global comparative analysis of DNA repair proteins and processes based upon the analysis of available complete genome sequences. We use a new form of analysis that combines genome sequence information and phylogenetic studies into a composite analysis we refer to as phylogenomics. We use this phylogenomic analysis to study the evolution of repair proteins and processes and to predict the repair phenotypes of those species for which we now know the complete genome sequence.
Figures
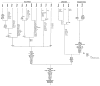
Figure 1. Demonstration of using evolutionary distribution patterns to trace gene gain and loss
An evolutionary tree of the relationships among some representatives of the bacteria, Archaea, and eukaryotes is shown. Presence of genes in these species is indicated by a colored box at the tip of the terminal branches of the tree. Gain and loss of the gene is inferred through parsimony reconstruction techniques. Within the bacterial part of the tree, we divide the species into major phyla but have collapsed the branches joining the different phyla to indicate that the relationships among these phyla are ambiguous.
Figure 2
Schematic diagram of an alignment of alkyltransferase genes.
Figure 3. Evolutionary gain and loss of DNA repair genes
The gain and loss of repair genes is traced onto an evolutionary tree of the species for which complete genome sequences were analyzed. Gain and loss were inferred by methods described in the main text. Origins of repair genes (+) are indicated on the branches while loss of genes (−) is indicated along side the branches. Gene duplication events are indicated by a “d” while possible lateral transfers are indicated by a “t”.
Similar articles
- lon incompatibility associated with mutations causing SOS induction: null uvrD alleles induce an SOS response in Escherichia coli.
SaiSree L, Reddy M, Gowrishankar J. SaiSree L, et al. J Bacteriol. 2000 Jun;182(11):3151-7. doi: 10.1128/JB.182.11.3151-3157.2000. J Bacteriol. 2000. PMID: 10809694 Free PMC article. - DNA repair in three dimensions.
Pearl LH, Savva R. Pearl LH, et al. Trends Biochem Sci. 1995 Oct;20(10):421-6. doi: 10.1016/s0968-0004(00)89092-1. Trends Biochem Sci. 1995. PMID: 8533156 Review. No abstract available. - Interspecies recombination and mismatch repair. Generation of mosaic genes and genomes.
Matic I, Taddei F, Radman M. Matic I, et al. Methods Mol Biol. 2000;152:149-57. doi: 10.1385/1-59259-068-3:149. Methods Mol Biol. 2000. PMID: 10957975 No abstract available. - Recombination is essential for viability of an Escherichia coli dam (DNA adenine methyltransferase) mutant.
Marinus MG. Marinus MG. J Bacteriol. 2000 Jan;182(2):463-8. doi: 10.1128/JB.182.2.463-468.2000. J Bacteriol. 2000. PMID: 10629194 Free PMC article. - Genetic analysis of recombination in prokaryotes.
Lloyd RG, Sharples GJ. Lloyd RG, et al. Curr Opin Genet Dev. 1992 Oct;2(5):683-90. doi: 10.1016/s0959-437x(05)80127-3. Curr Opin Genet Dev. 1992. PMID: 1458021 Review.
Cited by
- Paradoxical DNA repair and peroxide resistance gene conservation in Bacillus pumilus SAFR-032.
Gioia J, Yerrapragada S, Qin X, Jiang H, Igboeli OC, Muzny D, Dugan-Rocha S, Ding Y, Hawes A, Liu W, Perez L, Kovar C, Dinh H, Lee S, Nazareth L, Blyth P, Holder M, Buhay C, Tirumalai MR, Liu Y, Dasgupta I, Bokhetache L, Fujita M, Karouia F, Eswara Moorthy P, Siefert J, Uzman A, Buzumbo P, Verma A, Zwiya H, McWilliams BD, Olowu A, Clinkenbeard KD, Newcombe D, Golebiewski L, Petrosino JF, Nicholson WL, Fox GE, Venkateswaran K, Highlander SK, Weinstock GM. Gioia J, et al. PLoS One. 2007 Sep 26;2(9):e928. doi: 10.1371/journal.pone.0000928. PLoS One. 2007. PMID: 17895969 Free PMC article. - Helicobacter pylori mutants defective in RuvC Holliday junction resolvase display reduced macrophage survival and spontaneous clearance from the murine gastric mucosa.
Loughlin MF, Barnard FM, Jenkins D, Sharples GJ, Jenks PJ. Loughlin MF, et al. Infect Immun. 2003 Apr;71(4):2022-31. doi: 10.1128/IAI.71.4.2022-2031.2003. Infect Immun. 2003. PMID: 12654822 Free PMC article. - The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium.
Eisen JA, Nelson KE, Paulsen IT, Heidelberg JF, Wu M, Dodson RJ, Deboy R, Gwinn ML, Nelson WC, Haft DH, Hickey EK, Peterson JD, Durkin AS, Kolonay JL, Yang F, Holt I, Umayam LA, Mason T, Brenner M, Shea TP, Parksey D, Nierman WC, Feldblyum TV, Hansen CL, Craven MB, Radune D, Vamathevan J, Khouri H, White O, Gruber TM, Ketchum KA, Venter JC, Tettelin H, Bryant DA, Fraser CM. Eisen JA, et al. Proc Natl Acad Sci U S A. 2002 Jul 9;99(14):9509-14. doi: 10.1073/pnas.132181499. Epub 2002 Jul 1. Proc Natl Acad Sci U S A. 2002. PMID: 12093901 Free PMC article. - Identification of 5-formyluracil DNA glycosylase activity of human hNTH1 protein.
Miyabe I, Zhang QM, Kino K, Sugiyama H, Takao M, Yasui A, Yonei S. Miyabe I, et al. Nucleic Acids Res. 2002 Aug 1;30(15):3443-8. doi: 10.1093/nar/gkf460. Nucleic Acids Res. 2002. PMID: 12140329 Free PMC article. - Evolution of the methyl directed mismatch repair system in Escherichia coli.
Putnam CD. Putnam CD. DNA Repair (Amst). 2016 Feb;38:32-41. doi: 10.1016/j.dnarep.2015.11.016. Epub 2015 Dec 2. DNA Repair (Amst). 2016. PMID: 26698649 Free PMC article. Review.
References
- Eisen JA. Mechanistic basis of microsatellite instability. In: Goldstein DB, Schlotterer C, editors. Microsatellites: Evolution and Applications. Oxford University Press; Oxford: 1999. pp. 34–48.
- Labarére J. DNA replication and repair. In: Maniloff J, editor. Mycoplasmas: Molecular Biology And Pathogenesis. American Society For Microbiology; Washington, D. C: 1992. pp. 309–323.
- Dybvig K, Voelker LL. Molecular biology of mycoplasmas. Annu Rev Microbiol. 1996;50:25–57. - PubMed
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases