The HAND1 basic helix-loop-helix transcription factor regulates trophoblast differentiation via multiple mechanisms - PubMed (original) (raw)
The HAND1 basic helix-loop-helix transcription factor regulates trophoblast differentiation via multiple mechanisms
I C Scott et al. Mol Cell Biol. 2000 Jan.
Abstract
The basic helix-loop-helix (bHLH) transcription factor genes Hand1 and Mash2 are essential for placental development in mice. Hand1 promotes differentiation of trophoblast giant cells, whereas Mash2 is required for the maintenance of giant cell precursors, and its overexpression prevents giant cell differentiation. We found that Hand1 expression and Mash2 expression overlap in the ectoplacental cone and spongiotrophoblast, layers of the placenta that contain the giant cell precursors, indicating that the antagonistic activities of Hand1 and Mash2 must be coordinated. MASH2 and HAND1 both heterodimerize with E factors, bHLH proteins that are the DNA-binding partners for most class B bHLH factors and which are also expressed in the ectoplacental cone and spongiotrophoblast. In vitro, HAND1 could antagonize MASH2 function by competing for E-factor binding. However, the Hand1 mutant phenotype cannot be solely explained by ectopic activity of MASH2, as the Hand1 mutant phenotype was not altered by further mutation of Mash2. Interestingly, expression of E-factor genes (ITF2 and ALF1) was down-regulated in the trophoblast lineage prior to giant cell differentiation. Therefore, suppression of MASH2 function, required to allow giant cell differentiation, may occur in vivo by loss of its E-factor partner due to loss of its expression and/or competition from HAND1. In giant cells, where E-factor expression was not detected, HAND1 presumably associates with a different bHLH partner. This may account for the distinct functions of HAND1 in giant cells and their precursors. We conclude that development of the trophoblast lineage is regulated by the interacting functions of HAND1, MASH2, and their cofactors.
Figures
FIG. 1
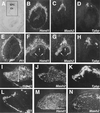
Hand1 and Mash2 have intersecting expression domains in trophoblast. (B to E) Serial sections of an E8.5 implantation site following RNA in situ hybridization with antisense probes for Hand1 (B), Mash2 (C), Tpbp (D), and Pl1 (E) shown in dark field. (A) Section B shown in light field. Hand1 and Mash2 expression overlaps in cells of the ectoplacental cone. (F to H) Increased magnification of panels B to D in boxed area depicted in panel A. Arrows indicate the chorion. (I to L) Expression of Hand1 (I), Mash2 (J), Tpbp (K), and Pl1 (L) in E10.5 placenta. (M and N) Expression of Hand1 (M) and Mash2 (N) in E12.5 placenta. (A to E) Panels I to N at ×50 magnification. Ch, chorion; EPC, ectoplacental cone; Lab, labyrinthine layer; Sp, spongiotrophoblast layer.
FIG. 2
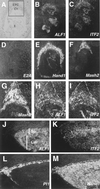
E-factor gene expression during trophoblast development. (A to F) Serial sections of an E8.5 implantation site hybridized with antisense probes specific to ALF1 (B), ITF2 (C), E2A (D), Hand1 (E), and Mash2 (F) shown in dark field. (A) Section B shown in light field. E-factor gene expression is undetectable in trophoblast giant cells. (G to I) Magnification of boxed area shown in panel A for Mash2 (G), ALF1 (H), and ITF2 (I) probes. Note that ALF1 and ITF2 expression does not extend to the periphery of the ectoplacental cone (dotted line). (J to M) Serial sections of E10.5 placenta hybridized with probes for ALF1 (J), ITF2 (K), Pl1 (L), and Hand1 (M). Hand1 is expressed in _Pl1_-positive giant cells (bounded by the dotted line). (A to G) Panels J to M at ×5 magnification. Ch, chorion; E, embryo; EPC, ectoplacental cone; Lab, labyrinthine layer; Sp, spongiotrophoblast layer.
FIG. 3
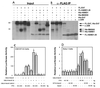
HAND1 can both homodimerize and heterodimerize with E factors. (A and B) Coimmunoprecipitation assays using in vitro-translated FL-E47 or FL-HAND1ΔN as bait for untagged HAND1, MASH2, E47, and c-Jun. (A) Proteins were mixed and subjected to SDS-PAGE; (B) mixed proteins were immunoprecipitated (IP) with anti-FLAG antibody M2, washed, and resolved by SDS-PAGE. (C and D) Two-hybrid assays. The pL8G5-Luc reporter, in which luciferase expression is driven by a minimal promoter and five copies of the GAL4 upstream activation sequence DNA-binding site, was used along with 100 ng of the indicated GAL4 fusion construct. Different superscripts indicate statistically significant differences (P < 0.05).
FIG. 4
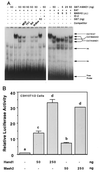
HAND1 inhibits MASH2 binding to MCK E boxes by titrating E factors. (A and C) Electrophoretic mobility shift assay using a labeled MCK E-box probe. For indicated reactions, in vitro-translated FL-E47 (2 μl), His-MASH2 (4 μl), His-ALF1 (2 μl), and His-ITF2 (2 μl) were added. For competition assays, a 200-fold excess of unlabeled oligonucleotide was used. ns, nonspecific complex. (B and D) Transfection assays using C3H10T1/2 cells and the p2E MCK-CAT reporter, in which a CAT gene is driven by a minimal promoter and two copies of an MCK E-box sequence. Different superscripts indicate statistically significant differences (P < 0.05).
FIG. 5
HAND1 and MASH2 bind and activate transcription from a Th1 E-box sequence as heterodimers with E factors. (A) Electrophoretic mobility shift assay using a labeled Th1 E-box probe. For indicated reactions, 2 and 4 μl of in vitro-translated FL-E47 and His-MASH2, respectively, were used. For competition assays, a 200-fold excess of unlabeled oligonucleotide was used. ns, nonspecific complex. (B) Transfection assays using C3H10T1/2 fibroblasts. The pL8E6-Luc reporter, in which luciferase expression is driven by a minimal promoter and six copies of the Th1 E box, was used. Different superscripts indicate statistically significant differences (P < 0.05).
FIG. 6
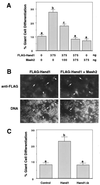
Mutation of the HAND1 basic domain abrogates its ability to promote trophoblast giant cell differentiation. (A and C) Rcho-1 trophoblast cells were transiently cotransfected with a lacZ marker and the indicated expression vector(s). β-Galactosidase-positive cells were scored for giant cell morphology 2 days posttransfection. Different superscripts indicate statistically significant differences (P < 0.05). (B) Immunofluorescent detection of FL-HAND1 expression in transfected cells. Arrows and arrowheads indicate FL-HAND1-negative and -positive cells, respectively.
FIG. 7
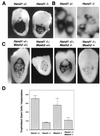
Decreased mural trophectoderm cell number and nuclear size in Hand1 mutants is independent of Mash2 function. (A) β-Galactosidase staining for the 6AD1βgeo allele in E8.5 Hand1 +/− and −/− implantation sites. (B) High-magnification view of panel C showing mural trophoblast cells along the lateral side of the implantation site. Pl1 transcript localization is perinuclear. (C) E8.5 implantation sites derived from crosses between Hand1 +/−; Mash2 +/− compound heterozygotes were bisected and subjected to whole-mount in situ hybridization using an antisense Pl1 probe. Shown is a low-magnification view of Pl1 expression in trophoblast giant cells in one-half of the implantation site. (D) _Pl1_-positive giant cells per conceptus were counted (three implantation sites per genotype).
FIG. 8
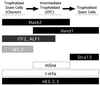
Distinct bHLH compartments in early trophoblast development. Shown is a summary of expression patterns of bHLH factors and modifiers in trophoblast at E8.5. EPC, ectoplacental cone. Boxes: black, bHLH factors; dark gray, E factors; light gray, HLH factors; white, non-HLH factors.
Similar articles
- Differential regulation of Hand1 homodimer and Hand1-E12 heterodimer activity by the cofactor FHL2.
Hill AA, Riley PR. Hill AA, et al. Mol Cell Biol. 2004 Nov;24(22):9835-47. doi: 10.1128/MCB.24.22.9835-9847.2004. Mol Cell Biol. 2004. PMID: 15509787 Free PMC article. - The Hand1, Stra13 and Gcm1 transcription factors override FGF signaling to promote terminal differentiation of trophoblast stem cells.
Hughes M, Dobric N, Scott IC, Su L, Starovic M, St-Pierre B, Egan SE, Kingdom JC, Cross JC. Hughes M, et al. Dev Biol. 2004 Jul 1;271(1):26-37. doi: 10.1016/j.ydbio.2004.03.029. Dev Biol. 2004. PMID: 15196947 - Mash2 acts cell autonomously in mouse spongiotrophoblast development.
Tanaka M, Gertsenstein M, Rossant J, Nagy A. Tanaka M, et al. Dev Biol. 1997 Oct 1;190(1):55-65. doi: 10.1006/dbio.1997.8685. Dev Biol. 1997. PMID: 9331331 - Transcription factors underlying the development and endocrine functions of the placenta.
Cross JC, Anson-Cartwright L, Scott IC. Cross JC, et al. Recent Prog Horm Res. 2002;57:221-34. doi: 10.1210/rp.57.1.221. Recent Prog Horm Res. 2002. PMID: 12017545 Review. - E protein function in lymphocyte development.
Quong MW, Romanow WJ, Murre C. Quong MW, et al. Annu Rev Immunol. 2002;20:301-22. doi: 10.1146/annurev.immunol.20.092501.162048. Epub 2001 Oct 4. Annu Rev Immunol. 2002. PMID: 11861605 Review.
Cited by
- Differential regulation of Hand1 homodimer and Hand1-E12 heterodimer activity by the cofactor FHL2.
Hill AA, Riley PR. Hill AA, et al. Mol Cell Biol. 2004 Nov;24(22):9835-47. doi: 10.1128/MCB.24.22.9835-9847.2004. Mol Cell Biol. 2004. PMID: 15509787 Free PMC article. - L3MBTL1 deficiency directs the differentiation of human embryonic stem cells toward trophectoderm.
Hoya-Arias R, Tomishima M, Perna F, Voza F, Nimer SD. Hoya-Arias R, et al. Stem Cells Dev. 2011 Nov;20(11):1889-900. doi: 10.1089/scd.2010.0437. Epub 2011 Apr 3. Stem Cells Dev. 2011. PMID: 21341991 Free PMC article. - Somatic donor cell type correlates with embryonic, but not extra-embryonic, gene expression in postimplantation cloned embryos.
Hirasawa R, Matoba S, Inoue K, Ogura A. Hirasawa R, et al. PLoS One. 2013 Oct 16;8(10):e76422. doi: 10.1371/journal.pone.0076422. eCollection 2013. PLoS One. 2013. PMID: 24146866 Free PMC article. - Sex Specification and Heterogeneity of Primordial Germ Cells in Mice.
Sakashita A, Kawabata Y, Jincho Y, Tajima S, Kumamoto S, Kobayashi H, Matsui Y, Kono T. Sakashita A, et al. PLoS One. 2015 Dec 23;10(12):e0144836. doi: 10.1371/journal.pone.0144836. eCollection 2015. PLoS One. 2015. PMID: 26700643 Free PMC article. - Human Long Noncoding RNA Regulation of Stem Cell Potency and Differentiation.
Lee S, Seo HH, Lee CY, Lee J, Shin S, Kim SW, Lim S, Hwang KC. Lee S, et al. Stem Cells Int. 2017;2017:6374504. doi: 10.1155/2017/6374504. Epub 2017 Aug 30. Stem Cells Int. 2017. PMID: 28951743 Free PMC article. Review.
References
- Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. - PubMed
- Blanar M A, Rutter W J. Interaction cloning: identification of a helix-loop-helix zipper protein that interacts with c-Fos. Science. 1992;256:1014–1018. - PubMed
- Calzonetti T, Stevenson L, Rossant J. A novel regulatory region is required for trophoblast-specific transcription in transgenic mice. Dev Biol. 1995;171:615–626. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases