The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc - PubMed (original) (raw)
The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc
S B McMahon et al. Mol Cell Biol. 2000 Jan.
Abstract
The c-Myc protein functions as a transcription factor to facilitate oncogenic transformation; however, the biochemical and genetic pathways leading to transformation remain undefined. We demonstrate here that the recently described c-Myc cofactor TRRAP recruits histone acetylase activity, which is catalyzed by the human GCN5 protein. Since c-Myc function is inhibited by recruitment of histone deacetylase activity through Mad family proteins, these opposing biochemical activities are likely to be responsible for the antagonistic biological effects of c-Myc and Mad on target genes and ultimately on cellular transformation.
Figures
FIG. 1
The c-Myc N terminus recruits a HAT. (A) FLAG epitope-tagged GAL4 DBD or a FLAG epitope-tagged GAL4 DBD–c-Myc (amino acids 1 to 262) fusion was used as an affinity matrix to isolate proteins from HeLa cell nuclear extracts. (B) Proteins recruited by each matrix were incubated with purified histones in the presence of radiolabeled acetyl-CoA. Following filter binding and washing, the degree of histone acetylation in each sample was quantitated by scintillation counting. Assays were performed in triplicate. Values for individual samples after subtraction of background are reported.
FIG. 2
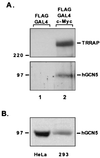
The c-Myc N terminus recruits the HAT hGCN5. (A) Proteins captured by the GAL4 DBD or GAL4 DBD–c-Myc fusion were resolved by SDS-PAGE (8% gel) and immunoblotted for either the c-Myc cofactor TRRAP (A) or the recently described mammalian HAT hGCN5 (B). Antibodies to hGCN5 were the generous gift from Nickolai Barlev and Shelley Berger. Numbers at left (in kilodaltons) indicate positions of size markers. (B) Western blotting for hGCN5 was performed with HeLa cell nuclear extracts and whole-cell lysates from 293 cells.
FIG. 3
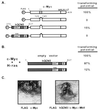
Myc family oncoproteins recruit hGCN5 in human cells. (A) 293 cells were lysed under nondenaturing conditions, and c-Myc–Max dimers were immunoprecipitated with antisera against Max (lanes 2 and 3). A parallel precipitation was performed with nonimmune rabbit serum (NRS) as a control (lane 1). Precipitated proteins were resolved by SDS-PAGE (8% gel) and subjected to immunoblotting for either c-Myc (lower panel), hGCN5 (middle panel), or TRRAP (upper panel). Numbers at the left (in kilodaltons) indicate positions of size markers. The position of the heavy-chain polypeptide from the precipitating antibody is indicated (Ig). (B) 293 cells were lysed and subjected to immunoprecipitation with either a control monoclonal antibody (ø) or a monoclonal antibody directed against c-Myc. Immunoprecipitates (i.p.) were blotted and probed for TRRAP (upper panel) and hGCN5 (lower panel). (C) 293 cells were transiently transfected with a cytomegalovirus-driven expression vector encoding FLAG epitope-tagged versions of either wild-type (wt) murine N-Myc (lane 2) or a mutant lacking amino acids 100 to 116 of the MbII domain (lane 3). Following transfection, cells were lysed as for panel A, immunoprecipitations were performed with anti-FLAG antibody; precipitates were resolved by SDS-PAGE and Western blotted for either the FLAG epitope (lower panel) or hGCN5 (upper panel). Mock transfected 293 cells served as a control (lane 1).
FIG. 4
The c-Myc oncoprotein associates with HAT activity in vivo. Immunoprecipitation of 293 cell lysates for either c-Myc, Max, or hGCN5 were subjected to an in vitro HAT assay in the presence of purified histones and radiolabeled acetyl-CoA. For each precipitation, lysate was prepared from a single confluent 15-cm-diameter dish of serum-stimulated 293 cells. Precipitations contained approximately 4 mg of total protein and were conducted in triplicate. Bars represent average values (± standard error).
FIG. 5
In vivo association of hGCN5 and TRRAP in the absence of c-Myc. (A) Aliquots of 293 cell lysates were either electrophoresed directly (lane 1) or subjected to immunoprecipitation with nonimmune rabbit serum (NRS; lane 2) or anti-hGCN5 (lane 3). Proteins were resolved by SDS-PAGE (8% gel) and immunoblotted for TRRAP (upper panel) or hGCN5 (lower panel). Numbers at the left (in kilodaltons) indicate positions of molecular weight markers. (B) 293 cell lysates were subjected to immunoprecipitation with antibodies specific for hGCN5 (lane 2), c-Myc (lane 4), or species-matched, nonimmune sera (lanes 1 and 3). Precipitated proteins were resolved by SDS-PAGE (8% gel) and immunoblotted for c-Myc. The position of the heavy-chain polypeptide from the precipitating antibody is indicated (Ig).
FIG. 6
Partial rescue of a nontransforming c-Myc mutant by direct fusion to hGCN5. (A) Primary rat embryo fibroblasts were transfected with expression vectors for an activated allele of p21H-ras and one of the c-Myc constructs schematized at the left. The c-Myc constructs encoded either wild-type mouse c-Myc or a deletion mutant of this protein lacking 17 amino acids from the MbII domain. In addition, two fusion proteins were generated between the c-Myc MbII domain deletion mutant and the catalytic domain of hGCN5 (amino acids 370 to 837). For the first of these fusions, wild-type hGCN5 sequences were fused to c-Myc ΔMbII. The second fusion contained three single amino acid substitutions within the hGCN5 HAT domain (as indicated by x's). These mutations have been shown previously to block both HAT activity and transcriptional activation when introduced into the corresponding residues of S. cerevisiae GCN5. The critical bromodomain of hGCN5 (labeled B) was also included in these fusion proteins. Transforming potential of each of these proteins was determined by examining cells approximately 2 weeks posttransfection. Transforming potential was estimated based on both the total number of transformed foci generated with a given protein and the size of the individual foci obtained. bHLH-LZ, basic helix-loop-helix leucine zipper. (B) A transformation assay was performed as for panel A except that full-length versions of hGCN5 were coexpressed with c-Myc. Both wild-type and catalytically inactive forms of hGCN5 were assayed for their effect on c-Myc mediated transformation. (C) Representative foci obtained by transfection of rat embryo fibroblasts with p21H-ras and either wild-type c-Myc (left) or the MbII mutant-hGCN5 fusion protein (right).
FIG. 7
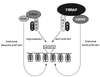
Model of the opposing biochemical functions of c-Myc and Mad. Mad family proteins dimerize with Max and repress the expression of c-myc and max target genes, thereby blocking c-Myc function. Mad-mediated repression requires the recruitment of a multiprotein complex containing histone deacetylases of the HDAC family. The demonstration here that c-Myc, through its essential cofactor TRRAP, recruits the mammalian HAT hGCN5 suggests a potential biochemical basis for antagonistic biological functions of c-Myc and Mad. In this model, c-Myc–Max heterodimers activate transcription of target genes by recruitment of TRRAP and a HAT such as hGCN5. HAT activity results in nucleosomal remodeling at target gene loci, allowing more efficient transcription. Following displacement of Myc-Max dimers from their cognate DNA recognition element, Mad-Max dimers recruit deacetylase activity to these sites, which in turn removes the acetyl groups from histones in nearby nucleosomes. This process facilitates chromatin condensation and consequently transcriptional repression.
Similar articles
- The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis.
Park J, Kunjibettu S, McMahon SB, Cole MD. Park J, et al. Genes Dev. 2001 Jul 1;15(13):1619-24. doi: 10.1101/gad.900101. Genes Dev. 2001. PMID: 11445536 Free PMC article. - MYC interacts with the human STAGA coactivator complex via multivalent contacts with the GCN5 and TRRAP subunits.
Zhang N, Ichikawa W, Faiola F, Lo SY, Liu X, Martinez E. Zhang N, et al. Biochim Biophys Acta. 2014 May;1839(5):395-405. doi: 10.1016/j.bbagrm.2014.03.017. Epub 2014 Apr 3. Biochim Biophys Acta. 2014. PMID: 24705139 Free PMC article. - c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation.
Liu X, Tesfai J, Evrard YA, Dent SY, Martinez E. Liu X, et al. J Biol Chem. 2003 May 30;278(22):20405-12. doi: 10.1074/jbc.M211795200. Epub 2003 Mar 26. J Biol Chem. 2003. PMID: 12660246 Free PMC article. - Repression by the Mad(Mxi1)-Sin3 complex.
Schreiber-Agus N, DePinho RA. Schreiber-Agus N, et al. Bioessays. 1998 Oct;20(10):808-18. doi: 10.1002/(SICI)1521-1878(199810)20:10<808::AID-BIES6>3.0.CO;2-U. Bioessays. 1998. PMID: 9819568 Review. - Transcriptional regulation. Flipping the Myc switch.
Bernards R. Bernards R. Curr Biol. 1995 Aug 1;5(8):859-61. doi: 10.1016/s0960-9822(95)00173-4. Curr Biol. 1995. PMID: 7583141 Review.
Cited by
- Conservation and diversity of the eukaryotic SAGA coactivator complex across kingdoms.
Chen YC, Dent SYR. Chen YC, et al. Epigenetics Chromatin. 2021 Jun 10;14(1):26. doi: 10.1186/s13072-021-00402-x. Epigenetics Chromatin. 2021. PMID: 34112237 Free PMC article. Review. - The tumor suppressor protein HBP1 is a novel c-myc-binding protein that negatively regulates c-myc transcriptional activity.
Escamilla-Powers JR, Daniel CJ, Farrell A, Taylor K, Zhang X, Byers S, Sears R. Escamilla-Powers JR, et al. J Biol Chem. 2010 Feb 12;285(7):4847-58. doi: 10.1074/jbc.M109.074856. Epub 2009 Dec 11. J Biol Chem. 2010. PMID: 20008325 Free PMC article. - Metastasis-associated protein 1 (MTA1) is an essential downstream effector of the c-MYC oncoprotein.
Zhang XY, DeSalle LM, Patel JH, Capobianco AJ, Yu D, Thomas-Tikhonenko A, McMahon SB. Zhang XY, et al. Proc Natl Acad Sci U S A. 2005 Sep 27;102(39):13968-73. doi: 10.1073/pnas.0502330102. Epub 2005 Sep 19. Proc Natl Acad Sci U S A. 2005. PMID: 16172399 Free PMC article. - Formation of a structurally-stable conformation by the intrinsically disordered MYC:TRRAP complex.
Feris EJ, Hinds JW, Cole MD. Feris EJ, et al. PLoS One. 2019 Dec 2;14(12):e0225784. doi: 10.1371/journal.pone.0225784. eCollection 2019. PLoS One. 2019. PMID: 31790487 Free PMC article. - A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism.
Nikiforov MA, Chandriani S, O'Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy JM, Cole MD. Nikiforov MA, et al. Mol Cell Biol. 2002 Aug;22(16):5793-800. doi: 10.1128/MCB.22.16.5793-5800.2002. Mol Cell Biol. 2002. PMID: 12138190 Free PMC article.
References
- Ayer D E, Kretzner L, Eisenman R N. Mad: a heterodimeric partner for max that antagonizes myc transcriptional activity. Cell. 1993;72:211–222. - PubMed
- Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. - PubMed
- Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
- Bouchard C, Staller P, Eilers M. Control of cell proliferation by Myc. Trends Cell Biol. 1998;8:202–206. - PubMed
- Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases