Targeted mRNA degradation by double-stranded RNA in vitro - PubMed (original) (raw)
Targeted mRNA degradation by double-stranded RNA in vitro
T Tuschl et al. Genes Dev. 1999.
Abstract
Double-stranded RNA (dsRNA) directs gene-specific, post-transcriptional silencing in many organisms, including vertebrates, and has provided a new tool for studying gene function. The biochemical mechanisms underlying this dsRNA interference (RNAi) are unknown. Here we report the development of a cell-free system from syncytial blastoderm Drosophila embryos that recapitulates many of the features of RNAi. The interference observed in this reaction is sequence specific, is promoted by dsRNA but not single-stranded RNA, functions by specific mRNA degradation, and requires a minimum length of dsRNA. Furthermore, preincubation of dsRNA potentiates its activity. These results demonstrate that RNAi can be mediated by sequence-specific processes in soluble reactions.
Figures
Figure 1
Reporter mRNAs and dsRNAs. (A) RNAs used in this study. Lengths and positions of the ssRNA, asRNA, and dsRNAs are shown as black bars relative to the _Rr_-Luc and _Pp_-Luc reporter mRNA sequences. Black rectangles indicate the two unrelated luciferase coding sequences, lines correspond to the 5′ and 3′ UTRs of the mRNAs. (B) Native gel electrophoresis of the individual Rr 501 nt and Pp 505 nt asRNAs and ssRNAs used to form the Rr and Pp dsRNAs.
Figure 2
Gene-specific interference by dsRNA in vitro. (A) Ratio of luciferase activities after targeting 50 p
m
_Pp_-Luc mRNA with 10 n
m
ssRNA, asRNA, or dsRNA from the 505-bp segment of the _Pp_-Luc gene. The data are the average values of seven trials ± standard deviation. Four independently prepared lysates were used. Luciferase activity was normalized to the buffer control and so a ratio equal to one indicates no gene-specific interference. (B) Ratio of luciferase activities after targeting 50 p
m
_Rr_-Luc mRNA with 10 n
m
ssRNA, asRNA, or dsRNA from the 501-bp segment of the _Rr_-Luc gene. The data are the average values of six trials ± standard deviation. A _Rr_-Luc/_Pp_-Luc ratio equal to one indicates no gene-specific interference.
Figure 2
Gene-specific interference by dsRNA in vitro. (A) Ratio of luciferase activities after targeting 50 p
m
_Pp_-Luc mRNA with 10 n
m
ssRNA, asRNA, or dsRNA from the 505-bp segment of the _Pp_-Luc gene. The data are the average values of seven trials ± standard deviation. Four independently prepared lysates were used. Luciferase activity was normalized to the buffer control and so a ratio equal to one indicates no gene-specific interference. (B) Ratio of luciferase activities after targeting 50 p
m
_Rr_-Luc mRNA with 10 n
m
ssRNA, asRNA, or dsRNA from the 501-bp segment of the _Rr_-Luc gene. The data are the average values of six trials ± standard deviation. A _Rr_-Luc/_Pp_-Luc ratio equal to one indicates no gene-specific interference.
Figure 3
Incubation in the Drosophila embryo lysate potentiates dsRNA for gene-specific interference. (A) Experimental strategy. The same dsRNAs used in Fig. 2 (or buffer) was serially preincubated with twofold dilutions in six successive reactions with Drosophila embryo lysate, then tested for its capacity to block mRNA expression. As a control, the same amount of dsRNA (10 n
m
) or buffer was diluted directly in buffer and incubated with _Pp_-Luc and _Rr_-Luc mRNAs and lysate. (B) Potentiation when targeting _Pp_-Luc mRNA. Black columns indicate the dsRNA or the buffer was serially preincubated; white columns correspond to a direct 32-fold dilution of the dsRNA. Values were normalized to those of the buffer controls. (C) Potentiation when targeting _Rr_-Luc mRNA. The corresponding buffer control is shown in B.
Figure 3
Incubation in the Drosophila embryo lysate potentiates dsRNA for gene-specific interference. (A) Experimental strategy. The same dsRNAs used in Fig. 2 (or buffer) was serially preincubated with twofold dilutions in six successive reactions with Drosophila embryo lysate, then tested for its capacity to block mRNA expression. As a control, the same amount of dsRNA (10 n
m
) or buffer was diluted directly in buffer and incubated with _Pp_-Luc and _Rr_-Luc mRNAs and lysate. (B) Potentiation when targeting _Pp_-Luc mRNA. Black columns indicate the dsRNA or the buffer was serially preincubated; white columns correspond to a direct 32-fold dilution of the dsRNA. Values were normalized to those of the buffer controls. (C) Potentiation when targeting _Rr_-Luc mRNA. The corresponding buffer control is shown in B.
Figure 3
Incubation in the Drosophila embryo lysate potentiates dsRNA for gene-specific interference. (A) Experimental strategy. The same dsRNAs used in Fig. 2 (or buffer) was serially preincubated with twofold dilutions in six successive reactions with Drosophila embryo lysate, then tested for its capacity to block mRNA expression. As a control, the same amount of dsRNA (10 n
m
) or buffer was diluted directly in buffer and incubated with _Pp_-Luc and _Rr_-Luc mRNAs and lysate. (B) Potentiation when targeting _Pp_-Luc mRNA. Black columns indicate the dsRNA or the buffer was serially preincubated; white columns correspond to a direct 32-fold dilution of the dsRNA. Values were normalized to those of the buffer controls. (C) Potentiation when targeting _Rr_-Luc mRNA. The corresponding buffer control is shown in B.
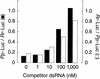
Figure 4
Effect of competitor dsRNA on gene-specific interference. Increasing concentrations of nanos dsRNA (508 bp) were added to reactions containing 5 n
m
dsRNA (the same dsRNAs used in Fig. 2) targeting _Pp_-Luc mRNA (black columns, left axis) or _Rr_-Luc mRNA (white columns, right axis). Each reaction contained both a target mRNA (_Pp_-Luc for the black columns, _Rr_-Luc for the white) and an unrelated control mRNA (_Rr_-Luc for the black columns, _Pp_-Luc for the white). Values were normalized to the buffer control (not shown). The reactions were incubated under standard conditions (see Materials and Methods).
Figure 5
Effect of dsRNA on mRNA stability. (A) Stability of 10 n
m
_Pp_-Luc mRNA or _Rr_-Luc mRNA incubated in lysate with either buffer or 505-bp _Pp_-dsRNA (10 n
m
). Samples were deproteinized after the indicated times and the 32P-radiolabeled mRNAs were then resolved by denaturing gel electrophoresis. The band marked with an asterisk likely results from radioactivity being swept ahead of the abundant ribosomal RNA in the lysate. (B) Quantitation of the data in A. (Circles) _Pp_-Luc mRNA; (boxes) _Rr_-Luc mRNA; (filled symbols) buffer incubation; (open symbols) incubation with _Pp_-dsRNA. (C) Stability of _Rr_-Luc mRNA incubated with _Rr_-dsRNA or _Pp_-dsRNA. (█) buffer; (□) _Pp_-dsRNA (10 n
m
); (○) _Rr_-dsRNA (10 n
m
). (D) Dependence on dsRNA length. The stability of the _Pp_-Luc mRNA was assessed after incubation in lysate in the presence of buffer or dsRNAs of different lengths. (█) Buffer; (○) 49-bp dsRNA (10 n
m
); (▿) 149-bp dsRNA (10 n
m
); (▵) 505-bp dsRNA (10 n
m
); (⋄) 997-bp dsRNA (10 n
m
). Reactions were incubated under standard conditions (see Materials and Methods).
References
- Baulcombe DC. Fast forward genetics based on virus-induced gene silencing. Curr Opin Plant Biol. 1999;2:109–113. - PubMed
- Carthew, R. 1999. www1.pitt.edu/∼carthew/manual/RNAi_Protocol.html
- Clemens M, Williams B. Inhibition of cell-free protein synthesis by pppA2′p5′A2′5′A: a novel oligonucleotide synthesized by interferon-treated L cell extracts. Cell. 1978;13:565–572. - PubMed
- Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases