Three-dimensional structure of poliovirus receptor bound to poliovirus - PubMed (original) (raw)
Three-dimensional structure of poliovirus receptor bound to poliovirus
D M Belnap et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Poliovirus initiates infection by binding to its cellular receptor (Pvr). We have studied this interaction by using cryoelectron microscopy to determine the structure, at 21-A resolution, of poliovirus complexed with a soluble form of its receptor (sPvr). This density map aided construction of a homology-based model of sPvr and, in conjunction with the known crystal structure of the virus, allowed delineation of the binding site. The virion does not change significantly in structure on binding sPvr in short incubations at 4 degrees C. We infer that the binding configuration visualized represents the initial interaction that is followed by structural changes in the virion as infection proceeds. sPvr is segmented into three well-defined Ig-like domains. The two domains closest to the virion (domains 1 and 2) are aligned and rigidly connected, whereas domain 3 diverges at an angle of approximately 60 degrees. Two nodules of density on domain 2 are identified as glycosylation sites. Domain 1 penetrates the "canyon" that surrounds the 5-fold protrusion on the capsid surface, and its binding site involves all three major capsid proteins. The inferred pattern of virus-sPvr interactions accounts for most mutations that affect the binding of Pvr to poliovirus.
Figures
Figure 1
(a) Cryomicrographs of poliovirus particles complexed with (Top) and without (Bottom) sPvr. Bar = 300 Å. Image reconstructions are shown of virion + sPvr [in stereo (b)] and, for comparison, of the virion (c) (from ref. 9). The two reconstructions were overlaid in d with the respective contour levels adjusted to clarify the interaction of sPvr with the virion. Bar = 100 Å. (e) Two views of a single sPvr molecule extracted from the difference map. Domain boundaries are marked. Bar = 25 Å.
Figure 2
(a) A central section through the virion–sPvr reconstruction, normal to a 2-fold symmetry axis. The boxed region is shown in b. High density is shown as dark. The right edge of the box coincides with a 5-fold symmetry axis. (b) The virion–sPvr (Top) and virion (Bottom, from ref. 9) maps were oriented as in a rotated progressively about the 5-fold symmetry axis. (Left to Right) The rotation is −18°, 0°, 6°, 12°, 24°, and 30°. In each case, the boxed portion of the central section was extracted. Arrows indicate sPvr-related density that bridges the canyon (second panel from right) and the solvent-level density corresponding to the tunnel (rightmost panel). Bars = 50 Å.
Figure 3
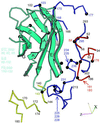
(a) “Road map” representations (36, 37) of poliovirus (Left) and rhinovirus-14 [Right; (33)]. The corresponding triangular area of the capsid surface, bounded by a 5-fold and two 3-fold icosahedral symmetry axes, is marked (Inset). The radial distances of surface residues from the virion center are color coded and contoured [see key (Top Right)]. Nomenclature: 3145, residue 145 of VP3. The receptor footprints are shown in white. (b) A ribbon diagram (38) of the sPvr model is flanked by two views (39) of a single sPvr molecule as portrayed in the cryoelectron microscopy density map (white cage), enclosing the model of the three sPvr domains, d1 (cyan), d2 (orange), and d3 (violet). Carbohydrates attached to d2 [to N188 (Left) and N237 (Right)] and possibly to d1 are shown in brown. Also shown are the capsid proteins VP1 (blue), VP2 (yellow), VP3 (red), and VP4 (green). The tunnel beneath the sPvr-binding site is evident (white arrows). “Pocket factor” is magenta. (c) The sPvr sequence is mapped onto secondary structural elements of the homology model. Asn residues thought to be glycosylated are marked with asterisks. (d) Ribbon diagram (38) showing the docking of the sPvr model onto the capsid surface. Same color conventions as in b. The axes allow this view to be related to Fig. 4. (e) Schematic diagram showing a possible binding configuration of poliovirus with intact membrane-bound Pvr.
Figure 4
Ribbon diagram (38) of the capsid-binding d1 domain of Pvr (cyan), with β-strands labeled, juxtaposed with segments of the capsid proteins with which it is inferred to interact. The viral segments are shown as tubes, with VP1, blue; VP2, yellow; and VP3, red. Residue numbers are provided as landmarks. Black balls and colored numbers denote amino acids implicated by genetic analysis in receptor binding. Similarly, Pvr residues shown by mutation to be important for virus binding are listed (Left). The axes allow this view to be related to Fig. 3_d_.
Similar articles
- Interaction of the poliovirus receptor with poliovirus.
He Y, Bowman VD, Mueller S, Bator CM, Bella J, Peng X, Baker TS, Wimmer E, Kuhn RJ, Rossmann MG. He Y, et al. Proc Natl Acad Sci U S A. 2000 Jan 4;97(1):79-84. doi: 10.1073/pnas.97.1.79. Proc Natl Acad Sci U S A. 2000. PMID: 10618374 Free PMC article. - Complexes of poliovirus serotypes with their common cellular receptor, CD155.
He Y, Mueller S, Chipman PR, Bator CM, Peng X, Bowman VD, Mukhopadhyay S, Wimmer E, Kuhn RJ, Rossmann MG. He Y, et al. J Virol. 2003 Apr;77(8):4827-35. doi: 10.1128/jvi.77.8.4827-4835.2003. J Virol. 2003. PMID: 12663789 Free PMC article. - Cryo-electron microscopy reconstruction of a poliovirus-receptor-membrane complex.
Bubeck D, Filman DJ, Hogle JM. Bubeck D, et al. Nat Struct Mol Biol. 2005 Jul;12(7):615-8. doi: 10.1038/nsmb955. Epub 2005 Jun 19. Nat Struct Mol Biol. 2005. PMID: 15965485 Free PMC article. - Early events in poliovirus infection: virus-receptor interactions.
Racaniello VR. Racaniello VR. Proc Natl Acad Sci U S A. 1996 Oct 15;93(21):11378-81. doi: 10.1073/pnas.93.21.11378. Proc Natl Acad Sci U S A. 1996. PMID: 8876143 Free PMC article. Review. - Poliovirus cell entry: common structural themes in viral cell entry pathways.
Hogle JM. Hogle JM. Annu Rev Microbiol. 2002;56:677-702. doi: 10.1146/annurev.micro.56.012302.160757. Epub 2002 Jan 30. Annu Rev Microbiol. 2002. PMID: 12142481 Free PMC article. Review.
Cited by
- Inhibition of coxsackie B virus infection by soluble forms of its receptors: binding affinities, altered particle formation, and competition with cellular receptors.
Goodfellow IG, Evans DJ, Blom AM, Kerrigan D, Miners JS, Morgan BP, Spiller OB. Goodfellow IG, et al. J Virol. 2005 Sep;79(18):12016-24. doi: 10.1128/JVI.79.18.12016-12024.2005. J Virol. 2005. PMID: 16140777 Free PMC article. - Antigenic evolution of vaccine-derived polioviruses: changes in individual epitopes and relative stability of the overall immunological properties.
Yakovenko ML, Cherkasova EA, Rezapkin GV, Ivanova OE, Ivanov AP, Eremeeva TP, Baykova OY, Chumakov KM, Agol VI. Yakovenko ML, et al. J Virol. 2006 Mar;80(6):2641-53. doi: 10.1128/JVI.80.6.2641-2653.2006. J Virol. 2006. PMID: 16501074 Free PMC article. - Structure of decay-accelerating factor bound to echovirus 7: a virus-receptor complex.
He Y, Lin F, Chipman PR, Bator CM, Baker TS, Shoham M, Kuhn RJ, Medof ME, Rossmann MG. He Y, et al. Proc Natl Acad Sci U S A. 2002 Aug 6;99(16):10325-9. doi: 10.1073/pnas.152161599. Epub 2002 Jul 15. Proc Natl Acad Sci U S A. 2002. PMID: 12119400 Free PMC article. - Interaction of coxsackievirus A21 with its cellular receptor, ICAM-1.
Xiao C, Bator CM, Bowman VD, Rieder E, He Y, Hébert B, Bella J, Baker TS, Wimmer E, Kuhn RJ, Rossmann MG. Xiao C, et al. J Virol. 2001 Mar;75(5):2444-51. doi: 10.1128/JVI.75.5.2444-2451.2001. J Virol. 2001. PMID: 11160747 Free PMC article. - Retrospective characterization of a vaccine-derived poliovirus type 1 isolate from sewage in Greece.
Dedepsidis E, Kyriakopoulou Z, Pliaka V, Kottaridi C, Bolanaki E, Levidiotou-Stefanou S, Komiotis D, Markoulatos P. Dedepsidis E, et al. Appl Environ Microbiol. 2007 Nov;73(21):6697-704. doi: 10.1128/AEM.00535-07. Epub 2007 Sep 7. Appl Environ Microbiol. 2007. PMID: 17827314 Free PMC article.
References
- Racaniello V R. Structure (London) 1996;4:769–773. - PubMed
- Mendelsohn C L, Wimmer E, Racaniello V R. Cell. 1989;56:855–865. - PubMed
- Geraghty R J, Krummenacher C, Cohen G H, Eisenberg R J, Spear P G. Science. 1998;280:1618–1620. - PubMed
- Warner M S, Geraghty R J, Martinez W M, Montgomery R I, Whitbeck J C, Xu R, Eisenberg R J, Cohen G H, Spear P G. Virology. 1998;246:179–189. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI20017/AI/NIAID NIH HHS/United States
- AI20566/AI/NIAID NIH HHS/United States
- R21 AI020566/AI/NIAID NIH HHS/United States
- R37 AI020566/AI/NIAID NIH HHS/United States
- R01 AI020566/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials