NMR solution structure of the human prion protein - PubMed (original) (raw)
NMR solution structure of the human prion protein
R Zahn et al. Proc Natl Acad Sci U S A. 2000.
Abstract
The NMR structures of the recombinant human prion protein, hPrP(23-230), and two C-terminal fragments, hPrP(90-230) and hPrP(121-230), include a globular domain extending from residues 125-228, for which a detailed structure was obtained, and an N-terminal flexibly disordered "tail." The globular domain contains three alpha-helices comprising the residues 144-154, 173-194, and 200-228 and a short anti-parallel beta-sheet comprising the residues 128-131 and 161-164. Within the globular domain, three polypeptide segments show increased structural disorder: i.e., a loop of residues 167-171, the residues 187-194 at the end of helix 2, and the residues 219-228 in the C-terminal part of helix 3. The local conformational state of the polypeptide segments 187-193 in helix 2 and 219-226 in helix 3 is measurably influenced by the length of the N-terminal tail, with the helical states being most highly populated in hPrP(23-230). When compared with the previously reported structures of the murine and Syrian hamster prion proteins, the length of helix 3 coincides more closely with that in the Syrian hamster protein whereas the disordered loop 167-171 is shared with murine PrP. These species variations of local structure are in a surface area of the cellular form of PrP that has previously been implicated in intermolecular interactions related both to the species barrier for infectious transmission of prion disease and to immune reactions.
Figures
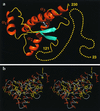
Figure 1
(a) Cartoon of the three-dimensional structure of the intact human prion protein, hPrP(23–230). The helices are orange, the β-strands cyan, the segments with nonregular secondary structure within the C-terminal domain yellow, and the flexibly disordered “tail” of residues 23–121 is represented by yellow dots. (b) Stereoview of an all-heavy atom presentation of the globular domain, with residues 125–228, in hPrP(23–230) in the same orientation as in a. The backbone is shown as a gray spline function through the Cα positions, hydrophobic side chains are yellow, and polar and charged side chains are orange. The figures were prepared with the program
molmol
(38).
Figure 2
Comparison of the mean NMR structures of the polypeptide segments with residues 125 to 228 in hPrP(23–230) (green), hPrP(90–230) (orange), and hPrP(121–230) (violet). A spline function was drawn through the Cα positions. The variable radius of the cylindrical rods is proportional to the mean global backbone displacement per residue (39), as evaluated after superposition for best fit of the atoms N, Cα, and C′ of the residues 125–228 in the 20 energy-minimized conformers used to represent the solution structure.
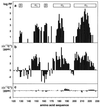
Figure 3
(a) Logarithmic plot of the amide proton exchange protection factors (PF) of residues 121–230 in hPrP(23–230) versus the sequence. Hydrogen–deuterium exchange was measured at 20°C in 99.9% D2O containing 10 mM sodium acetate and 0.05% sodium azide at pH 4.5. (b) 13Cα chemical shift differences, Δδ(13Cα), between hPrP(23–230) and the random coil shifts (51). (c) Δδ(13Cα) between hPrP(23–230) and hPrP(121–230) at pH 4.5 (Δδ = δ[hPrP(23–230)] − δ[hPrP(121–230)]). The locations of the regular secondary structure elements are given in a.
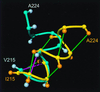
Figure 4
Comparison of helix 3 in hPrP(121–230) and mPrP(121–231), where the backbone of the polypeptide segment 215–224 is represented as a spline function drawn through the Cα positions. The figure results from a global superposition of the two proteins for best fit of the backbone atoms of the residues 144–154, 175–193, and 200–219, which correspond to the α-helices in mPrP. The following color code was used: yellow and orange, backbone and Cβ atoms of hPrP(121–230), respectively; cyan and light blue, backbone and Cβ atoms of mPrP(121–231), respectively; green, _d_αβ(i, i + 3) and _d_αN (i, i + 3) NOE distance constraints observed in hPrP; magenta, same types of NOE constraints observed for mPrP (11).
Similar articles
- NMR structure of the human doppel protein.
Lührs T, Riek R, Güntert P, Wüthrich K. Lührs T, et al. J Mol Biol. 2003 Mar 7;326(5):1549-57. doi: 10.1016/s0022-2836(02)01471-7. J Mol Biol. 2003. PMID: 12595265 - NMR structure of the bovine prion protein.
López Garcia F, Zahn R, Riek R, Wüthrich K. López Garcia F, et al. Proc Natl Acad Sci U S A. 2000 Jul 18;97(15):8334-9. doi: 10.1073/pnas.97.15.8334. Proc Natl Acad Sci U S A. 2000. PMID: 10899999 Free PMC article. - NMR structures of three single-residue variants of the human prion protein.
Calzolai L, Lysek DA, Guntert P, von Schroetter C, Riek R, Zahn R, Wüthrich K. Calzolai L, et al. Proc Natl Acad Sci U S A. 2000 Jul 18;97(15):8340-5. doi: 10.1073/pnas.97.15.8340. Proc Natl Acad Sci U S A. 2000. PMID: 10900000 Free PMC article. - Influence of pH on NMR structure and stability of the human prion protein globular domain.
Calzolai L, Zahn R. Calzolai L, et al. J Biol Chem. 2003 Sep 12;278(37):35592-6. doi: 10.1074/jbc.M303005200. Epub 2003 Jun 25. J Biol Chem. 2003. PMID: 12826672 - The prion protein: Structural features and related toxic peptides.
Ronga L, Tizzano B, Palladino P, Ragone R, Urso E, Maffia M, Ruvo M, Benedetti E, Rossi F. Ronga L, et al. Chem Biol Drug Des. 2006 Sep;68(3):139-47. doi: 10.1111/j.1747-0285.2006.00427.x. Chem Biol Drug Des. 2006. PMID: 17062011 Review.
Cited by
- Annotation of proteins of unknown function: initial enzyme results.
McKay T, Hart K, Horn A, Kessler H, Dodge G, Bardhi K, Bardhi K, Mills JL, Bernstein HJ, Craig PA. McKay T, et al. J Struct Funct Genomics. 2015 Mar;16(1):43-54. doi: 10.1007/s10969-015-9194-5. Epub 2015 Jan 29. J Struct Funct Genomics. 2015. PMID: 25630330 Free PMC article. - Guinea Pig Prion Protein Supports Rapid Propagation of Bovine Spongiform Encephalopathy and Variant Creutzfeldt-Jakob Disease Prions.
Watts JC, Giles K, Saltzberg DJ, Dugger BN, Patel S, Oehler A, Bhardwaj S, Sali A, Prusiner SB. Watts JC, et al. J Virol. 2016 Oct 14;90(21):9558-9569. doi: 10.1128/JVI.01106-16. Print 2016 Nov 1. J Virol. 2016. PMID: 27440899 Free PMC article. - Multimodal small-molecule screening for human prion protein binders.
Reidenbach AG, Mesleh MF, Casalena D, Vallabh SM, Dahlin JL, Leed AJ, Chan AI, Usanov DL, Yehl JB, Lemke CT, Campbell AJ, Shah RN, Shrestha OK, Sacher JR, Rangel VL, Moroco JA, Sathappa M, Nonato MC, Nguyen KT, Wright SK, Liu DR, Wagner FF, Kaushik VK, Auld DS, Schreiber SL, Minikel EV. Reidenbach AG, et al. J Biol Chem. 2020 Sep 25;295(39):13516-13531. doi: 10.1074/jbc.RA120.014905. Epub 2020 Jul 28. J Biol Chem. 2020. PMID: 32723867 Free PMC article. - Unfolding Mechanism and Fibril Formation Propensity of Human Prion Protein in the Presence of Molecular Crowding Agents.
Madheswaran M, Ventserova N, D'Abrosca G, Salzano G, Celauro L, Cazzaniga FA, Isernia C, Malgieri G, Moda F, Russo L, Legname G, Fattorusso R. Madheswaran M, et al. Int J Mol Sci. 2024 Sep 13;25(18):9916. doi: 10.3390/ijms25189916. Int J Mol Sci. 2024. PMID: 39337404 Free PMC article. - Use of proteinase K nonspecific digestion for selective and comprehensive identification of interpeptide cross-links: application to prion proteins.
Petrotchenko EV, Serpa JJ, Hardie DB, Berjanskii M, Suriyamongkol BP, Wishart DS, Borchers CH. Petrotchenko EV, et al. Mol Cell Proteomics. 2012 Jul;11(7):M111.013524. doi: 10.1074/mcp.M111.013524. Epub 2012 Mar 21. Mol Cell Proteomics. 2012. PMID: 22438564 Free PMC article.
References
- Weissmann C. FEBS Lett. 1996;389:3–11. - PubMed
- Alper T, Cramp W A, Haig D A, Clarke M C. Nature (London) 1967;214:764–766. - PubMed
- Griffith J S. Nature (London) 1967;215:1043–1044. - PubMed
- Prusiner S B. Trends Biochem Sci. 1996;21:482–487. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials