Analysis of receptor signaling pathways by mass spectrometry: identification of vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors - PubMed (original) (raw)
Analysis of receptor signaling pathways by mass spectrometry: identification of vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors
A Pandey et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Oligomerization of receptor protein tyrosine kinases such as the epidermal growth factor receptor (EGFR) by their cognate ligands leads to activation of the receptor. Transphosphorylation of the receptor subunits is followed by the recruitment of signaling molecules containing src homology 2 (SH2) or phosphotyrosine interaction domains (PID). Additionally, several cytoplasmic proteins that may or may not associate with the receptor undergo tyrosine phosphorylation. To identify several components of the EGFR signaling pathway in a single step, we have immunoprecipitated molecules that are tyrosine phosphorylated in response to EGF and analyzed them by one-dimensional gel electrophoresis followed by mass spectrometry. Combining matrix-assisted laser desorption/ionization (MALDI) and nanoelectrospray tandem mass spectrometry (MS/MS) led to the identification of nine signaling molecules, seven of which had previously been implicated in EGFR signaling. Several of these molecules were identified from low femtomole levels of protein loaded onto the gel. We identified Vav-2, a recently discovered guanosine nucleotide exchange factor that is expressed ubiquitously, as a substrate of the EGFR. We demonstrate that Vav-2 is phosphorylated on tyrosine residues in response to EGF and associates with the EGFR in vivo. Binding of Vav-2 to the EGFR is mediated by the SH2 domain of Vav-2. In keeping with its ubiquitous expression, Vav-2 seems to be a general signaling molecule, since it also associates with the platelet-derived growth factor (PDGF) receptor and undergoes tyrosine phosphorylation in fibroblasts upon PDGF stimulation. The strategy suggested here can be used for routine identification of downstream components of cell surface receptors in mammalian cells.
Figures
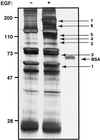
Figure 1
EGF-induced tyrosine phosphorylation in HeLa cells. Serum-deprived HeLa S3 cells (5 × 109) were either left untreated or treated with 1 μg/ml EGF for 5 min. Cleared cell lysates were immunoprecipitated with a mixture of monoclonal anti-phosphotyrosine antibodies, washed, and resolved by SDS/PAGE. The gel was then silver-stained. Molecular mass markers in kDa are indicated as well as 150 fmol of BSA that was loaded onto the same gel. The arrows with numbers indicate the positions of the bands that were excised for enzymatic digestion by trypsin and subsequent mass spectrometric analysis.
Figure 2
Use of mass spectrometry to identify Vav-2 and to resolve protein mixtures. Proteins separated by SDS/PAGE shown in Fig. 1 were subjected to in-gel digestion by trypsin and analyzed by mass spectrometry. (A) Protein band 6 was analyzed by MALDI. The tryptic peptides from the digest indicate the presence of several proteins in this mixture. Filled squares correspond to peptides derived from EGFR, T refers to trypsin autolysis products, and M denotes matrix ions. The peaks marked with stars denote a novel protein. Peaks marked with filled circles correspond to Eps15 (confirmed by MS/MS). (B) Protein band 4 was analyzed by MALDI. The tryptic peptides from this band showed the presence of peptides corresponding to Vav-2 (labeled V) and Hrs (shown by open circles). T refers to trypsin autolysis products and M denotes matrix ions. (C) MS spectrum from nanoelectrospray MS/MS analysis of the peptides from the sample analyzed by MALDI in B. V shows a peak corresponding to Vav-2, and T refers to trypsin autolysis products and their sodium adducts. (Inset) Isotopic resolution of a doubly charged peptide corresponding to Vav-2. (D) Fragmentation of the doubly charged peptide ([M+2H]2) shown in C (m/z = 794.48) by MS/MS. The Y" series of ions (C-terminal fragments) that are produced due to fragmentation are shown as well as one from the B series (N-terminal fragments; b2). The sequence of the peptide derived from this spectrum is shown at the top of the panel.
Figure 3
Vav-2 is tyrosine phosphorylated by EGF treatment. HeLa cells were either left untreated or treated with EGF for 5 min, and lysates were immunoprecipitated (IP) with anti-Vav-2 antibody as indicated. Washed immunoprecipitates were resolved by SDS/PAGE, transferred onto nitrocellulose, and Western blotted with an anti-phosphotyrosine antibody (A). The position of tyrosine-phosphorylated Vav-2 is indicated by an arrow. B shows equal loading of Vav-2 from a parallel experiment.
Figure 4
Vav-2 associates with EGFR by means of its SH2 domain. (A) HeLa cells either were left untreated or were treated with EGF, and lysates were immunoprecipitated with anti-Vav-2 antibody as indicated. Washed immunoprecipitates were resolved by SDS/PAGE, transferred onto nitrocellulose, and Western blotted with an anti-phosphotyrosine antibody. Coimmunoprecipitated EGFR is indicated by an arrow. (B) HeLa cells either were left untreated or were treated with EGF, and lysates were incubated with 10 μg of GST alone, GST-SH2, or GST-SH3 (C-terminal) fusion proteins bound to glutathione-agarose beads for 2 hr at 4°C. The beads were then washed and boiled in sample buffer. The samples were resolved by SDS/PAGE, transferred onto nitrocellulose, and Western blotted with an anti-phosphotyrosine antibody to detect the EGFR.
Figure 5
Vav-2 is a substrate of the PDGF receptor in fibroblasts. (A) NIH 3T3 fibroblasts either were left untreated or were treated with PDGF, and lysates were immunoprecipitated with anti-Vav-2 antibody as indicated. Washed immunoprecipitates were resolved by SDS/PAGE, transferred onto nitrocellulose, and Western blotted with an anti-phosphotyrosine antibody. Coimmunoprecipitated PDGFR and Vav-2 are indicated by arrows. (B) Anti-Vav-2 immunoprecipitates from a parallel experiment to show equal loading of Vav-2.
Similar articles
- Vav family proteins couple to diverse cell surface receptors.
Moores SL, Selfors LM, Fredericks J, Breit T, Fujikawa K, Alt FW, Brugge JS, Swat W. Moores SL, et al. Mol Cell Biol. 2000 Sep;20(17):6364-73. doi: 10.1128/MCB.20.17.6364-6373.2000. Mol Cell Biol. 2000. PMID: 10938113 Free PMC article. - The FRK/RAK-SHB signaling cascade: a versatile signal-transduction pathway that regulates cell survival, differentiation and proliferation.
Annerén C, Lindholm CK, Kriz V, Welsh M. Annerén C, et al. Curr Mol Med. 2003 Jun;3(4):313-24. doi: 10.2174/1566524033479744. Curr Mol Med. 2003. PMID: 12776987 Review. - Molecular mechanisms of SH2- and PTB-domain-containing proteins in receptor tyrosine kinase signaling.
Wagner MJ, Stacey MM, Liu BA, Pawson T. Wagner MJ, et al. Cold Spring Harb Perspect Biol. 2013 Dec 1;5(12):a008987. doi: 10.1101/cshperspect.a008987. Cold Spring Harb Perspect Biol. 2013. PMID: 24296166 Free PMC article. Review.
Cited by
- Proteomic Interrogation in Cancer Biomarker.
Kang UB. Kang UB. Adv Exp Med Biol. 2021;1187:305-322. doi: 10.1007/978-981-32-9620-6_15. Adv Exp Med Biol. 2021. PMID: 33983585 - Low-bias phosphopeptide enrichment from scarce samples using plastic antibodies.
Chen J, Shinde S, Koch MH, Eisenacher M, Galozzi S, Lerari T, Barkovits K, Subedi P, Krüger R, Kuhlmann K, Sellergren B, Helling S, Marcus K. Chen J, et al. Sci Rep. 2015 Jul 1;5:11438. doi: 10.1038/srep11438. Sci Rep. 2015. PMID: 26126808 Free PMC article. - Strong cation exchange chromatography in analysis of posttranslational modifications: innovations and perspectives.
Edelmann MJ. Edelmann MJ. J Biomed Biotechnol. 2011;2011:936508. doi: 10.1155/2011/936508. Epub 2011 Nov 17. J Biomed Biotechnol. 2011. PMID: 22174558 Free PMC article. Review. - Phosphoproteomics: new insights into cellular signaling.
Mumby M, Brekken D. Mumby M, et al. Genome Biol. 2005;6(9):230. doi: 10.1186/gb-2005-6-9-230. Epub 2005 Aug 17. Genome Biol. 2005. PMID: 16168091 Free PMC article. Review. - In-depth analyses of kinase-dependent tyrosine phosphoproteomes based on metal ion-functionalized soluble nanopolymers.
Iliuk AB, Martin VA, Alicie BM, Geahlen RL, Tao WA. Iliuk AB, et al. Mol Cell Proteomics. 2010 Oct;9(10):2162-72. doi: 10.1074/mcp.M110.000091. Epub 2010 Jun 17. Mol Cell Proteomics. 2010. PMID: 20562096 Free PMC article.
References
- Cross M, Dexter M. Cell. 1991;64:271–280. - PubMed
- Ullrich A, Schlessinger J. Cell. 1990;61:203–212. - PubMed
- Pawson T. Nature (London) 1995;373:573–580. - PubMed
- Nishibe S, Wahl M I, Hernandez-Sotomayor S M, Tonks N K, Rhee S G, Carpenter G. Science. 1990;250:1253–1256. - PubMed
- Kim H K, Kim J W, Zilberstein A, Margolis B, Kim J G, Schlessinger J, Rhee S G. Cell. 1991;65:435–441. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous