Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter - PubMed (original) (raw)
Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter
L Wang et al. Proc Natl Acad Sci U S A. 2000.
Abstract
The Epstein-Barr virus (EBV) nuclear protein 2 (EBNA2) and herpes simplex virion protein 16 (VP16) acidic domains that mediate transcriptional activation now are found to have affinity for p300, CBP, and PCAF histone acetyltransferases (HATs). Transcriptionally inactive point mutations in these domains lack affinity for p300, CBP, or PCAF. P300 and CBP copurify with the principal HAT activities that bind to EBNA2 or VP16 acidic domains through velocity sedimentation and anion-exchange chromatography. EBNA2 binds to both the N- and C-terminal domains of p300 and coimmune-precipitates from transfected 293T cells with p300. In EBV-infected Akata Burkitt's tumor cells that do not express the EBV encoded oncoproteins EBNA2 or LMP1, p300 expression enhances the ability of EBNA2 to up-regulate LMP1 expression. Through its intrinsic HAT activity, PCAF can further potentiate the p300 effect. In 293 T cells, P300 and CBP (but not PCAF) can also coactivate transcription mediated by the EBNA2 or VP16 acidic domains and HAT-negative mutants of p300 have partial activity. Thus, the EBNA2 and VP16 acidic domains can utilize the intrinsic HAT or scaffolding properties of p300 to activate transcription.
Figures
Figure 1
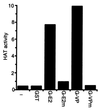
HAT activity from BJAB lysates binds specifically to the EBNA2 and VP16 acidic domains. The wild-type EBNA2 (E2) or VP16 (VP) or their null point mutant (E2m and VPm) acidic domains were fused to GST (G), and glutathione-Sepharose beads coated with fusion protein were used in pull downs with extracts from BJAB B cell lymphoma cells. The presence of HAT activity in proteins bound to beads was determined by liquid HAT assay. HAT activity was shown as 103 × count per minute. Results from one representative experiment of five similar experiments are shown.
Figure 2
Purification of GST-EBNA2-associated HAT activity. BJAB cell extracts were incubated with GST-EBNA2 beads. The HAT activity was eluted from the beads with 0.2 M NaCl and loaded onto 10–30% sucrose gradient. Fractions containing HAT activity were pooled, applied to anion-exchange column, and eluted with a linear 50- to 300-mM NaCl gradient. Protein fractions from sucrose gradient (A) and UNO Q column (B) were separated by SDS/PAGE and identified by Western blot with anti-CBP/p300 antibody (Lower). The HAT activity of these fractions also was measured (Upper) and shown in 104 × count per minute. Fraction numbers are indicated just below the Upper parts of A and B.
Figure 3
The EBNA2 and VP16 acidic domains specifically bind p300, CBP, and PCAF. BJAB cell extracts were incubated with glutathione-Sepharose beads loaded with wild-type GST-EBNA2 or GST-VP16 acidic domains or with the null point mutants. The amount of p300, CBP, or PCAF that bound to the beads was assayed by SDS/PAGE followed by Western blot with anti-CBP/p300 antibody or with anti-PCAF antibody.
Figure 4
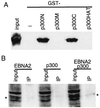
EBNA2 interacts with p300 in vitro and in vivo. (A) EBNA2 interacts with amino- and carboxyl-terminal of p300 in vitro. GST fusions to the p300 amino-terminal (amino acids 1–585, p300N), middle (amino acids 766–1571, p300M), carboxyl-terminal (amino acids 1572–2370, p300C), or HAT (amino acids 1195–1701, p300HAT) domains were expressed in E. coli, purified by using glutathione-Sepharose beads, and incubated with His-tagged full-length wild-type EBNA2 protein that has also been purified from E. coli. EBNA2 binding to GST-p300 beads was assayed by Western blot with an EBNA2-specific mAb (PE2) after SDS/PAGE of proteins that bound to the beads. The amount of protein used in the binding assays was five times that shown as input. (B) EBNA2 coimmune-precipitates with p300 from 293 T cells overexpressing both proteins. EBNA2, flag-tagged p300, or both were transfected into 293 T cells. Cell extracts were incubated with M2 beads to immune-precipitate flag-tagged p300, and EBNA2 was detected by Western blot with PE2 mAb. P300 was precipitated from 10 times the amount of lysate shown as input.
Figure 5
P300 coactivates LMP1 expression mediated by EBNA2 in Akata EBV-infected but EBNA2-negative cells, and PCAF potentiates the activation. Akata cells were transfected with 10 μg of SG5-EBNA2, 10 μg of SG5-p300, 20 μg of pCI-PCAF, or 20 μg of HAT-negative PCAF mutants Δ579–608 and Δ609–624 protein expression vectors. Empty vector was used to balance the transfections to 40 μg of total DNA when necessary. Whole-cell extracts were Western-blotted with the S12 anti-LMP1 mAb.
Figure 6
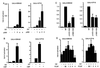
The effect of p300, CBP, or PCAF overexpression on GAL4-EBNA2 (GE) or GAL4-VP16 (GV) acidic domain-mediated transactivation of a GAL4-dependent promoter in 293 T cells. Various amounts of p300 expression vector (A) or 3.5 μg of CBP expression vector (B) was transfected into 293 T cell with 0.2 μg of GAL4-EBNA2 or 0.01 μg of GAL4-VP16 expression plasmids and 0.5 μg of reporter plasmid. The amount of cotransfected p300 DNA is indicated (in μg) in the figure. The fold activation induced by p300/CBP is relative to that induced by GAL4-EBNA2 or GAL4-VP16 alone. In C, the effect of 4 μg of p300 expression vector on coexpressed GAL4-EBNA2 acidic domain-mediated transactivation is compared with that of p300 HAT-negative mutants p300Δ1472–1522 or Δ1603–1653 in 293 T cells. The extent of cotransactivation by p300 mutants is shown relative to that by wild-type p300. In D, the effect of PCAF on GAL4-EBNA2- or GAL4-VP16-mediated activation of a GAL4-responsive promoter is assayed as described in A above. For all experiments in this figure, vector DNA was used to balance the different amounts of DNA used in different transfections. Luciferase activity was adjusted to the transfection efficiency represented by β-galactosidase activity. The results shown are the average of four to five independent experiments, with error bars representing SE.
Similar articles
- PCAF interacts with tax and stimulates tax transactivation in a histone acetyltransferase-independent manner.
Jiang H, Lu H, Schiltz RL, Pise-Masison CA, Ogryzko VV, Nakatani Y, Brady JN. Jiang H, et al. Mol Cell Biol. 1999 Dec;19(12):8136-45. doi: 10.1128/MCB.19.12.8136. Mol Cell Biol. 1999. PMID: 10567539 Free PMC article. - The amino-terminal C/H1 domain of CREB binding protein mediates zta transcriptional activation of latent Epstein-Barr virus.
Zerby D, Chen CJ, Poon E, Lee D, Shiekhattar R, Lieberman PM. Zerby D, et al. Mol Cell Biol. 1999 Mar;19(3):1617-26. doi: 10.1128/MCB.19.3.1617. Mol Cell Biol. 1999. PMID: 10022850 Free PMC article. - The histone acetyltransferase activity of PCAF cooperates with the brahma/SWI2-related protein BRG-1 in the activation of the enhancer A of the MHC class I promoter.
Brockmann D, Lehmkühler O, Schmücker U, Esche H. Brockmann D, et al. Gene. 2001 Oct 17;277(1-2):111-20. doi: 10.1016/s0378-1119(01)00703-x. Gene. 2001. PMID: 11602348 - CBP and p300: HATs for different occasions.
Kalkhoven E. Kalkhoven E. Biochem Pharmacol. 2004 Sep 15;68(6):1145-55. doi: 10.1016/j.bcp.2004.03.045. Biochem Pharmacol. 2004. PMID: 15313412 Review. - Acetylation of general transcription factors by histone acetyltransferases.
Imhof A, Yang XJ, Ogryzko VV, Nakatani Y, Wolffe AP, Ge H. Imhof A, et al. Curr Biol. 1997 Sep 1;7(9):689-92. doi: 10.1016/s0960-9822(06)00296-x. Curr Biol. 1997. PMID: 9285713 Review.
Cited by
- Epigenetic mechanisms in virus-induced tumorigenesis.
Poreba E, Broniarczyk JK, Gozdzicka-Jozefiak A. Poreba E, et al. Clin Epigenetics. 2011 Aug;2(2):233-47. doi: 10.1007/s13148-011-0026-6. Epub 2011 Mar 23. Clin Epigenetics. 2011. PMID: 22704339 Free PMC article. - Notch1IC partially replaces EBNA2 function in B cells immortalized by Epstein-Barr virus.
Gordadze AV, Peng R, Tan J, Liu G, Sutton R, Kempkes B, Bornkamm GW, Ling PD. Gordadze AV, et al. J Virol. 2001 Jul;75(13):5899-912. doi: 10.1128/JVI.75.13.5899-5912.2001. J Virol. 2001. PMID: 11390591 Free PMC article. - Histone acetyltransferase-deficient p300 mutants in diffuse large B cell lymphoma have altered transcriptional regulatory activities and are required for optimal cell growth.
Haery L, Lugo-Picó JG, Henry RA, Andrews AJ, Gilmore TD. Haery L, et al. Mol Cancer. 2014 Feb 15;13:29. doi: 10.1186/1476-4598-13-29. Mol Cancer. 2014. PMID: 24529102 Free PMC article. - Molecular Basis of Epstein-Barr Virus Latency Establishment and Lytic Reactivation.
Murata T, Sugimoto A, Inagaki T, Yanagi Y, Watanabe T, Sato Y, Kimura H. Murata T, et al. Viruses. 2021 Nov 23;13(12):2344. doi: 10.3390/v13122344. Viruses. 2021. PMID: 34960613 Free PMC article. Review. - Phase separation of Epstein-Barr virus EBNA2 protein reorganizes chromatin topology for epigenetic regulation.
Yang Y, Ye X, Dai R, Li Z, Zhang Y, Xue W, Zhu Y, Feng D, Qin L, Wang X, Lei B, Liao S, Hao B. Yang Y, et al. Commun Biol. 2021 Aug 16;4(1):967. doi: 10.1038/s42003-021-02501-7. Commun Biol. 2021. PMID: 34400762 Free PMC article.
References
- Alfieri C, Birkenbach M, Kieff E. Virology. 1991;181:595–608. - PubMed
- Henkel T, Ling P D, Hayward S D, Peterson M G. Science. 1994;265:92–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous