CREB activation induces adipogenesis in 3T3-L1 cells - PubMed (original) (raw)
CREB activation induces adipogenesis in 3T3-L1 cells
J E Reusch et al. Mol Cell Biol. 2000 Feb.
Abstract
Obesity is the result of numerous, interacting behavioral, physiological, and biochemical factors. One increasingly important factor is the generation of additional fat cells, or adipocytes, in response to excess feeding and/or large increases in body fat composition. The generation of new adipocytes is controlled by several "adipocyte-specific" transcription factors that regulate preadipocyte proliferation and adipogenesis. Generally these adipocyte-specific factors are expressed only following the induction of adipogenesis. The transcription factor(s) that are involved in initiating adipocyte differentiation have not been identified. Here we demonstrate that the transcription factor, CREB, is constitutively expressed in preadipocytes and throughout the differentiation process and that CREB is stimulated by conventional differentiation-inducing agents such as insulin, dexamethasone, and dibutyryl cAMP. Stably transfected 3T3-L1 preadipocytes were generated in which we could induce the expression of either a constitutively active CREB (VP16-CREB) or a dominant-negative CREB (KCREB). Inducible expression of VP16-CREB alone was sufficient to initiate adipogenesis as determined by triacylglycerol storage, cell morphology, and the expression of two adipocyte marker genes, peroxisome proliferator activated receptor gamma 2, and fatty acid binding protein. Alternatively, KCREB alone blocked adipogenesis in cells treated with conventional differentiation-inducing agents. These data indicate that activation of CREB was necessary and sufficient to induce adipogenesis. Finally, CREB was shown to bind to putative CRE sequences in the promoters of several adipocyte-specific genes. These data firmly establish CREB as a primary regulator of adipogenesis and suggest that CREB may play similar roles in other cells and tissues.
Figures
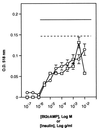
FIG. 1
Insulin or Bt2cAMP alone are sufficient to induce adipogenesis in 3T3-L1 cells. 3T3-L1 fibroblasts were passaged as described in Materials and Methods. The cells were treated with the indicated final concentrations of either Bt2cAMP (○) or insulin (□) for 48 h in high-glucose medium. The cells were then refed every 2 days for 10 days with high-glucose medium containing 10 μg of insulin per ml. On day 10 cells were fixed and stained with Oil Red O. Oil Red O was extracted from the cells with 200 μl of isopropanol and measured at 518 nm. Levels of Oil Red O staining were corrected for nonspecific binding levels of stain to untreated cells. For comparison, levels of Oil Red O staining in cells treated with either 10 μg of insulin per ml and 0.3 mM Bt2cAMP (dotted line) or else 10 μg of insulin per ml, 0.3 mM Bt2cAMP, and 1 μM dexamethasone (solid line) are also shown.
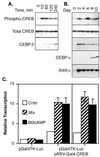
FIG. 2
CREB is expressed before and during adipogenesis, and differentiation-inducing agents stimulate CREB phoshorylation and transcriptional activity. (A) 3T3-L1 preadipocytes were grown to confluency as described in Materials and Methods. The cells were refed with complete growth medium containing 1 μg of insulin per ml, 1 μM dexamethasone, and 0.3 mM Bt2cAMP for the times indicated above each lane. Approximately 25 μg of cell lysate protein from each sample was separated on 10% acrylamide–SDS gels and transferred to nitrocellulose membranes. Duplicate membranes were subjected to Western analysis by using antibodies specific for Ser133 phosphorylated CREB (Phospho-CREB), total CREB, or CEBPβ protein as indicated. (B) Preadipocytes were grown to confluency and then refed with medium containing insulin, dexamethasone, and Bt2cAMP for 48 h. The cells were then refed every 2 days with medium containing 1 μg of insulin per ml. Cell lysates were prepared on the days indicated above each lane, and 25 μg of lysate protein from each sample was separated on 10% polyacrylamide–SDS gels and transferred to nitrocellulose membranes. Individual membranes were probed with antibodies specific for phospho-CREB, total CREB, CEBPα and -β, and RXRα as indicated. (C) 3T3-L1 fibroblasts and adipocytes were grown as described in Materials and Methods. Cells were transfected with the plasmid, pGal4TK-Luc, alone or with the plasmid pRSV-Gal4-CREB by using Superfect transfection reagent. The plasmids are described in the main text. At 24 h after transfection, the cells were treated with 0.5 mM Bt2cAMP alone or with a mixture of 1 μg of insulin per ml, 1 μM dexamethasone, and 0.5 mM Bt2cAMP for 4 h. The control cells received no treatment. Luciferase levels were measured in cell lysates as an index of transcription from the Gal4-TK promoter. Levels of transcription are shown relative to levels measured in untreated control cells transfected with pGal4TK-Luc alone.
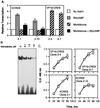
FIG. 3
Time course of VP16-CREB or KCREB expression in stably transfected 3T3-L1 cells after muristerone induction. 3T3-L1 cells stably transfected with the plasmid, pVgRXR, and either pIND-VP16-CREB or pIND-KCREB were generated as described in Materials and Methods. Individual clones inducibly expressing VP16-CREB, designated 2-4 and 9-7, and clones inducibly expressing KCREB, designated 2-1 and 2-10, were isolated. The expression of VP16-CREB or KCREB was monitored in these clonal cell lines versus time after treatment with muristerone at a final concentration of 10 μM. At 20 h, duplicate wells of cells were refed with medium lacking muristerone (levels indicated by dashed lines) for comparison to cells in medium with muristerone (solid lines). Levels of VP16-CREB and KCREB were measured by separating 25 μg of protein from lysates prepared at the times shown on 10% acrylamide–SDS gels. Proteins were transferred to nitrocellulose membranes subsequently probed with antibodies to VP16 (for VP16-CREB) or CREB (for KCREB). Since the CREB antibody detected both KCREB and endogenous CREB proteins, levels of KCREB expression were corrected for endogenous CREB levels measured in untreated cells (not shown). The optical densities of the bands on the blots was determined by using Scan Analysis Software. A representative blot for each protein is displayed to the right of the graphs.
FIG. 4
Effect of VP16-CREB or KCREB on CRE-dependent gene transcription and 3T3-L1 proliferation. (A) Stably transfected 3T3-L1 clonal cell lines inducibly expressing VP16-CREB (clones 2-4 and 9-7) or KCREB (clones 2-1 and 2-10) were transfected with the plasmid, −109 pPC-Luc, which contains a CREB-responsive portion of PEPCK gene promoter linked to a luciferase reporter gene by using Superfect reagent. The cells were cotransfected with the plasmid pSV-βGal. The following day the cells were treated with 0.3 mM Bt2cAMP, 10 μM muristerone, or both agents together as indicated. After 4 h cell lysates were prepared, and the luciferase activity was measured as an index of transcriptional activity. Levels are shown relative to the levels of luciferase activity in untreated, control cells (No Add'n). (B) 3T3-L1 cells stably transfected with pIND-KCREB (clone 2-10) were stimulated with the indicated final concentrations of muristerone for 20 h. Nuclear extracts were prepared, and electrophoretic mobility shift assays were performed with a 32P-labeled oligonucleotide containing the consensus CRE sequence (TGACGTCA). Reactions were separated on nondenaturing 6% polyacrylamide gels and exposed to film. The figure shows a representative autoradiograph. (C) Stably transfected 3T3-L1 clonal cell lines inducibly expressing VP16-CREB (clones 2-4 and 9-7) or KCREB (clones 2-1 and 2-10) were transferred to duplicate wells of 96-well plates (5,000 cells/well). After 24 h, one well was treated with 10 μg of muristerone (squares, solid line), and the remaining well was left untreated (circles, dotted line). Cell numbers were determined in each well with the Cell Titer 96 AQ reagent system at 24, 48, 72, and 96 h after plating of the cells. Values are shown relative to levels measured in wells containing untreated, control cells at the 24-h time point for each cell line and are the averages of three assays.
FIG. 5
VP16-CREB stimulates and KCREB inhibits adipogenesis in 3T3-L1 cells as determined by triacylglycerol storage. 3T3-L1 preadipocyte cell lines inducibly expressing VP16-CREB (clones 2-4 and 9-7) or KCREB (clones 2-1 and 2-10) or control cells (stably transfected with the plasmids, pVgRXR and pIND-LacZ) were grown to confluence as described in Materials and Methods. The cells were treated with the reagents indicated above each column of photographs. Cells treated with differentiation mixture received 10 μg of insulin per ml, 1 μM dexamethasone, and 0.3 mM Bt2cAMP for 48 h and then were refed every 2 days with conventional medium containing 10 μg of insulin per ml. Muristerone was added to medium at a final concentration of 10 μM for the entire 10-day differentiation period. After 10 days in culture, the cells were stained with Oil Red O to visualize triacylglycerol vesicles and then counterstained with hematoxylin. The photographs show cells on day 10 of each treatment.
FIG. 6
VP16 CREB stimulates and KCREB inhibits adipogenesis in 3T3-L1 cells as determined by expression of adipocyte-specific genes. Control and VP16-CREB- and KCREB-expressing cell lines were grown and treated as described in the legend to Fig. 5. On days 0 and 10 of the experiment, whole-cell lysates and total RNA was prepared from duplicate wells of cells. Approximately 25 μg of protein in the cell lysates was separated on 10% polyacrylamide–SDS gels and transferred to nitrocellulose blots. The blots were probed with a polyclonal antibody to PPARγ2, and the specific PPARγ2 band is indicated by an asterisk in the top row of blots. Likewise, 10 μg of total RNA was separated on 1% denaturing agarose gels and transferred to nitrocellulose blots. The blots were probed with an alkaline phosphatase-conjugated single-stranded oligonucleotide to FABP (aP2/422).
FIG. 7
Parameters of adipogenesis in VP16-CREB- or KCREB-expressing 3T3-L1 cells. Mixtures of clonal cell lines expressing VP16-CREB (clone 2-4 plus clone 9-7) or KCREB (clone 2-1 plus clone 2-10) were passaged as described in Materials and Methods as indicated. Cultures of these mixed cell populations were treated with the agents listed above each photograph. The numbers in parentheses indicates the days on which the agents were included in the growth medium for the cells. After 9 days in culture, the cells were fixed and stained with Oil Red O and counterstained with hematoxylin.
FIG. 8
CREB binds putative CRE sequences in the promoters of several adipocyte-specific genes. (A) The promoter regions of several adipocyte-specific genes were visually inspected for the presence of putative CRE sequences. Potential CREs present in these promoters are indicated by the box-enclosed regions which surround the nucleotides homologous to those in the consensus CRE sequences shown at the top of the figure. (B) Next, 20-bp double-stranded oligonucleotides, end labeled with [γ-32P]ATP and polynucleotide kinase, were incubated with purified, recombinant CREB protein as described in Materials and Methods. The reactions were separated on nondenaturing, 6% polyacrylamide gels and exposed to Kodak X-ARomat film. The figure shows a representative autoradiogram of the free (bottom) and CREB-bound complexes in comparison to reactions performed with a nonspecific (NS) oligonucleotide lacking a CRE sequence. (C) Next, 5 μg of nuclear extract protein prepared from 3T3-L1 fibroblasts was incubated with the indicated, labeled oligonucleotides either in the absence (−) or presence (+) of CREB-specific antibody. The reactions were separated on polyacrylamide gels as described above and exposed to film. The figure shows a representative autoradiogram of unbound and protein-bound oligonucleotides. (D) A total of 5 μg of nuclear extract protein prepared from 3T3-L1 untreated (−) fibroblasts or cells treated with muristerone to induce KCREB expression (+) was incubated with the indicated, labeled oligonucleotides. The reactions were separated on polyacrylamide gels as described above and exposed to film. The figure shows a representative autoradiogram of unbound and protein-bound oligonucleotides.
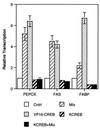
FIG. 9
CREB regulates transcription from adipocyte-specific gene promoters. Control or VP16-CREB- or KCREB-expressing 3T3-L1 fibroblasts were transfected with plasmids containing the full-length promoters of the PEPCK, FABP, or FAS genes linked to luciferase. The cells were cotransfected with the internal control plasmid, pRSV-βGal. The following day the cells were treated with muristerone to induce either VP16-CREB or KCREB expression as indicated and/or with the conventional differentiation mixture of 10 μg of insulin per ml, 1 μM dexamethasone, and 3 mM Bt2cAMP for 4 h. Cell lysates were then prepared, and luciferase activity was measured as an index of transcriptional activity. Levels of transcription are shown relative to levels measured in untreated cells for each promoter tested and were then corrected for transfection efficiency.
Similar articles
- Negative regulation of peroxisome proliferator-activated receptor-gamma gene expression contributes to the antiadipogenic effects of tumor necrosis factor-alpha.
Zhang B, Berger J, Hu E, Szalkowski D, White-Carrington S, Spiegelman BM, Moller DE. Zhang B, et al. Mol Endocrinol. 1996 Nov;10(11):1457-66. doi: 10.1210/mend.10.11.8923470. Mol Endocrinol. 1996. PMID: 8923470 - Overexpression and ribozyme-mediated targeting of transcriptional coactivators CREB-binding protein and p300 revealed their indispensable roles in adipocyte differentiation through the regulation of peroxisome proliferator-activated receptor gamma.
Takahashi N, Kawada T, Yamamoto T, Goto T, Taimatsu A, Aoki N, Kawasaki H, Taira K, Yokoyama KK, Kamei Y, Fushiki T. Takahashi N, et al. J Biol Chem. 2002 May 10;277(19):16906-12. doi: 10.1074/jbc.M200585200. Epub 2002 Mar 7. J Biol Chem. 2002. PMID: 11884404 - Expression of peroxisome proliferator-activated receptor PPARdelta promotes induction of PPARgamma and adipocyte differentiation in 3T3C2 fibroblasts.
Bastie C, Holst D, Gaillard D, Jehl-Pietri C, Grimaldi PA. Bastie C, et al. J Biol Chem. 1999 Jul 30;274(31):21920-5. doi: 10.1074/jbc.274.31.21920. J Biol Chem. 1999. PMID: 10419513 - A novel 3T3-L1 preadipocyte variant that expresses PPARgamma2 and RXRalpha but does not undergo differentiation.
Baillie RA, Sha X, Thuillier P, Clarke SD. Baillie RA, et al. J Lipid Res. 1998 Oct;39(10):2048-53. J Lipid Res. 1998. PMID: 9788251 - YM268 increases the glucose uptake, cell differentiation, and mRNA expression of glucose transporter in 3T3-L1 adipocytes.
Shimaya A, Kurosaki E, Shioduka K, Nakano R, Shibasaki M, Shikama H. Shimaya A, et al. Horm Metab Res. 1998 Sep;30(9):543-8. doi: 10.1055/s-2007-978929. Horm Metab Res. 1998. PMID: 9808320
Cited by
- Neddylation and Its Target Cullin 3 Are Essential for Adipocyte Differentiation.
Zhou H, Patel V, Rice R, Lee R, Kim HW, Weintraub NL, Su H, Chen W. Zhou H, et al. Cells. 2024 Oct 5;13(19):1654. doi: 10.3390/cells13191654. Cells. 2024. PMID: 39404417 Free PMC article. - Adipogenic Gene Expression in Gilthead Sea Bream Mesenchymal Stem Cells from Different Origin.
Salmerón C, Riera-Heredia N, Gutiérrez J, Navarro I, Capilla E. Salmerón C, et al. Front Endocrinol (Lausanne). 2016 Aug 22;7:113. doi: 10.3389/fendo.2016.00113. eCollection 2016. Front Endocrinol (Lausanne). 2016. PMID: 27597840 Free PMC article. - Alterations in chromatin accessibility during osteoblast and adipocyte differentiation in human mesenchymal stem cells.
Liu J, Gan L, Ma B, He S, Wu P, Li H, Xiong J. Liu J, et al. BMC Med Genomics. 2022 Jan 31;15(1):17. doi: 10.1186/s12920-022-01168-1. BMC Med Genomics. 2022. PMID: 35101056 Free PMC article. - Identification of a novel integral plasma membrane protein induced during adipocyte differentiation.
Albrektsen T, Richter HE, Clausen JT, Fleckner J. Albrektsen T, et al. Biochem J. 2001 Oct 15;359(Pt 2):393-402. doi: 10.1042/0264-6021:3590393. Biochem J. 2001. PMID: 11583587 Free PMC article. - Antiobesity effect of Kaempferia parviflora accompanied by inhibition of lipogenesis and stimulation of lipolysis.
Park SH, Park J, Lee M, Kim J, Eun S, Jun W, Kim OK, Lee J. Park SH, et al. Food Nutr Res. 2023 Jul 3;67. doi: 10.29219/fnr.v67.9374. eCollection 2023. Food Nutr Res. 2023. PMID: 37441513 Free PMC article.
References
- Abdel-Hafiz H A-M, Heasley L E, Kyriakis J M, Avruch J, Kroll D J, Johnson G L, Hoeffler J P. Activating transcription factor-2 DNA-binding activity is stimulated by phosphoryation catalyzed by p42 and p54 microtubule-associated protein kinases. Mol Endocrinol. 1992;6:2079–2089. - PubMed
- Abraham S, Johnson C L. Prevalence of severe obesity in adults in the United States. Am J Clin Nutr. 1980;33:364–369. - PubMed
- Amato S F, Nakajima K, Hirano T, Chiles T C. Transcriptional regulation of the junB gene in B lymphocytes: role of protein kinase A and a membrane Ig-regulated protein phosphatase. J Immunol. 1997;159:4676–4685. - PubMed
- Anonymous. Build study, 1979. Washington, D.C.: Society of Actuaries and Association of Life Insurance Medical Directors of America; 1980.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources