The rfaE gene from Escherichia coli encodes a bifunctional protein involved in biosynthesis of the lipopolysaccharide core precursor ADP-L-glycero-D-manno-heptose - PubMed (original) (raw)
The rfaE gene from Escherichia coli encodes a bifunctional protein involved in biosynthesis of the lipopolysaccharide core precursor ADP-L-glycero-D-manno-heptose
M A Valvano et al. J Bacteriol. 2000 Jan.
Abstract
The intermediate steps in the biosynthesis of the ADP-L-glycero-D-manno-heptose precursor of inner core lipopolysaccharide (LPS) are not yet elucidated. We isolated a mini-Tn10 insertion that confers a heptoseless LPS phenotype in the chromosome of Escherichia coli K-12. The mutation was in a gene homologous to the previously reported rfaE gene from Haemophilus influenzae. The E. coli rfaE gene was cloned into an expression vector, and an in vitro transcription-translation experiment revealed a polypeptide of approximately 55 kDa in mass. Comparisons of the predicted amino acid sequence with other proteins in the database showed the presence of two clearly separate domains. Domain I (amino acids 1 to 318) shared structural features with members of the ribokinase family, while Domain II (amino acids 344 to 477) had conserved features of the cytidylyltransferase superfamily that includes the aut gene product of Ralstonia eutrophus. Each domain was expressed individually, demonstrating that only Domain I could complement the rfaE::Tn10 mutation in E. coli, as well as the rfaE543 mutation of Salmonella enterica SL1102. DNA sequencing of the rfaE543 gene revealed that Domain I had one amino acid substitution and a 12-bp in-frame deletion resulting in the loss of four amino acids, while Domain II remained intact. We also demonstrated that the aut::Tn5 mutation in R. eutrophus is associated with heptoseless LPS, and this phenotype was restored following the introduction of a plasmid expressing the E. coli Domain II. Thus, both domains of rfaE are functionally different and genetically separable confirming that the encoded protein is bifunctional. We propose that Domain I is involved in the synthesis of D-glycero-D-manno-heptose 1-phosphate, whereas Domain II catalyzes the ADP transfer to form ADP-D-glycero-D-manno-heptose.
Figures
FIG. 1
Pathway for the synthesis of ADP-
l
-_glycero_-
d
-_manno_-heptose as proposed by Eidels and Osborn (15, 16). gmhA and gmhD are the only genes whose functions have been established biochemically (7, 10).
FIG. 2
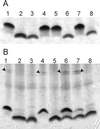
Analysis of LPS in E. coli and R. eutrophus. (A) LPS profiles of E. coli strains. Lanes: 1, W1485 (wild type); 2, KLC2926 (rfaE::Tn_10_); 3, χ711 (gmhA); 4, KLC2926(pTLS1); 5, KLC2926(pMAV51); 6, KLC2926(pMAV47); 7, KLC2926(pCM200); 8, KLC2926(pCM202). (B) LPS profiles of R. eutrophus strains. Lanes: 1, H16 (wild type); 2, HB3 (aut::Tn_5_); 3, E. coli χ711 (gmhA); 4, HB3(pMB10); 5, HB3(pMAV52); 6, HB3(pMAV53); 7, HB3(pCM204); 8, HB3(pCM205). Arrows indicate the bands corresponding to polymeric O antigen. In both cases, cell lysates were separated by Tricine-SDS-PAGE and stained with silver.
FIG. 3
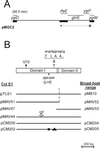
(A) Partial map of the chromosomal region cloned into pMGC2 that corresponds to the E. coli K-12 genetic map. Thick bars represent regions that were sequenced. The arrow indicates the direction of transcription of the putative ygiF-glnE-rfaE operon. (B) Map of the rfaE gene. Boxes correspond to the Domain I and Domain II regions. The positions of the various mutations are indicated: V70, the position of the Tn_10_ insertion in KLC2926 after the codon encoding valine 70 of the RfaE protein; ΔTLAA, the location of the 12-bp deletion in SL1102 rfaE and the corresponding amino acids; G→E, the glycine-to-glutamic acid substitution due to a base pair change (GGA to GAA). The partial maps show the various plasmids. The ColE1 vector was pEX1, and these plasmids were used in the experiments involving E. coli and S. enterica. The broad-host-range vector was pME6000, and these plasmids were used in the experiments involving R. eutrophus. The position and orientation (arrow) of the insertion of a cat cassette within the E. coli rfaE gene in pMAV44 is shown. pCM200 and pCM204 correspond to the cloned wild-type rfaE gene of S. enterica SL1027. pCM202 and pCM205 correspond to the cloned mutant rfaE543 gene of S. enterica SL1102. The box and triangle in pCM202/pCM205 denote the positions of the amino acid substitution and deletion, respectively.
FIG. 4
Amino acid sequencing alignment of Domain I of RfaE proteins from E. coli (RFAE_ECDI), S. enterica serovar Typhi (RFAE_TYPHI), S. enterica serovar Typhimurium strains SL11027 (RFAE_SL1027) and SL1102 (RFAE_SL1102), and E. coli ribokinase (RBSK_ECOLI). The alignment was produced with CLUSTAL W. Identical residues in all five proteins are boxed in black. ∗, residues that are also conserved in other members of the ribokinase family (45); r, a, and h, residues in the ribokinase that bind ribose (r) or ADP (a) via hydrogen bonds or that make van der Waals contacts with ribose and/or ADP (h) (for more details on the structure of ribokinase see reference 45). The arrow indicates the substitution of glycine (boxed in grey) by glutamic acid (bold) in the SL1102 RfaE mutant of SL1102. ▵, the deletion of four amino acids in the SL1102 RfaE mutant.
FIG. 5
Comparison of the known structure of the tyrosyl tRNA synthetase TyrTS (1tya_E) with Domain II. Secondary structure elements in TyrTS (6) are shown below the sequence (black bar, helix; gray bar, strand). The numbers indicate the helices and strands placed around the substrate binding site (according to reference 4). ● and ◊, conserved residues that line the ATP binding pocket. The flexible loop KFGKT, also involved in catalysis, and its corresponding segment in Domain II, STTNI (conserved in all members of cytidylyltransferases), are also boxed.
FIG. 6
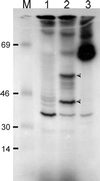
Autoradiograph showing rfaE expression by in vitro transcription-translation. Proteins were labeled with [14C]leucine as indicated by the supplier (Amersham) and separated by SDS-PAGE. M, [14C]methylated molecular weight markers (bovine serum albumin [69 kDa], ovalbumin [46 kDa], carbonic anhydrase [30 kDa], and lysozyme [14 kDa]). Lanes: 1, pMAV44; 2, pTLS1; 3, Kit's positive control. Arrowheads indicate unique polypeptides of ca. 55 and 38 kDa.
Similar articles
- Functional analysis of the glycero-manno-heptose 7-phosphate kinase domain from the bifunctional HldE protein, which is involved in ADP-L-glycero-D-manno-heptose biosynthesis.
McArthur F, Andersson CE, Loutet S, Mowbray SL, Valvano MA. McArthur F, et al. J Bacteriol. 2005 Aug;187(15):5292-300. doi: 10.1128/JB.187.15.5292-5300.2005. J Bacteriol. 2005. PMID: 16030223 Free PMC article. - Molecular cloning and functional expression of the rfaE gene required for lipopolysaccharide biosynthesis in Salmonella typhimurium.
Jin UH, Chung TW, Lee YC, Ha SD, Kim CH. Jin UH, et al. Glycoconj J. 2001 Oct;18(10):779-87. doi: 10.1023/a:1021103501626. Glycoconj J. 2001. PMID: 12441667 - ADP-heptose: A new innate immune modulator.
Hu X, Yang C, Wang PG, Zhang GL. Hu X, et al. Carbohydr Res. 2019 Feb 1;473:123-128. doi: 10.1016/j.carres.2018.12.011. Epub 2018 Dec 19. Carbohydr Res. 2019. PMID: 30684847 Review. - A Bittersweet Kiss of Gram-Negative Bacteria: The Role of ADP-Heptose in the Pathogenesis of Infection.
Sidor K, Skirecki T. Sidor K, et al. Microorganisms. 2023 May 17;11(5):1316. doi: 10.3390/microorganisms11051316. Microorganisms. 2023. PMID: 37317291 Free PMC article. Review.
Cited by
- Complex Multilevel Control of Hemolysin Production by Uropathogenic Escherichia coli.
Nhu NTK, Phan MD, Forde BM, Murthy AMV, Peters KM, Day CJ, Poole J, Kidd TJ, Welch RA, Jennings MP, Ulett GC, Sweet MJ, Beatson SA, Schembri MA. Nhu NTK, et al. mBio. 2019 Oct 1;10(5):e02248-19. doi: 10.1128/mBio.02248-19. mBio. 2019. PMID: 31575773 Free PMC article. - Large-scale analysis of the meningococcus genome by gene disruption: resistance to complement-mediated lysis.
Geoffroy MC, Floquet S, Métais A, Nassif X, Pelicic V. Geoffroy MC, et al. Genome Res. 2003 Mar;13(3):391-8. doi: 10.1101/gr.664303. Genome Res. 2003. PMID: 12618369 Free PMC article. - Functional analysis of the glycero-manno-heptose 7-phosphate kinase domain from the bifunctional HldE protein, which is involved in ADP-L-glycero-D-manno-heptose biosynthesis.
McArthur F, Andersson CE, Loutet S, Mowbray SL, Valvano MA. McArthur F, et al. J Bacteriol. 2005 Aug;187(15):5292-300. doi: 10.1128/JB.187.15.5292-5300.2005. J Bacteriol. 2005. PMID: 16030223 Free PMC article. - Identification of Genes Involved in Biogenesis of Outer Membrane Vesicles (OMVs) in Salmonella enterica Serovar Typhi.
Nevermann J, Silva A, Otero C, Oyarzún DP, Barrera B, Gil F, Calderón IL, Fuentes JA. Nevermann J, et al. Front Microbiol. 2019 Feb 4;10:104. doi: 10.3389/fmicb.2019.00104. eCollection 2019. Front Microbiol. 2019. PMID: 30778340 Free PMC article. - Comparative Evaluation of the Antimicrobial Activity of Different Antimicrobial Peptides against a Range of Pathogenic Bacteria.
Ebbensgaard A, Mordhorst H, Overgaard MT, Nielsen CG, Aarestrup FM, Hansen EB. Ebbensgaard A, et al. PLoS One. 2015 Dec 11;10(12):e0144611. doi: 10.1371/journal.pone.0144611. eCollection 2015. PLoS One. 2015. PMID: 26656394 Free PMC article.
References
- Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. - PubMed
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique EcoRI sites for selection of EcoRI generated recombinant DNA molecules. Gene. 1978;4:121–136. - PubMed
- Bork P, Holm L, Koonin E V, Sander C. The cytidylyltransferase superfamily: identification of the nucleotide-binding site and fold prediction. Proteins. 1995;22:259–266. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases