Multiple domains in caveolin-1 control its intracellular traffic - PubMed (original) (raw)
Multiple domains in caveolin-1 control its intracellular traffic
T Machleidt et al. J Cell Biol. 2000.
Abstract
Caveolin-1 is an integral membrane protein of caveolae that is thought to play an important role in both the traffic of cholesterol to caveolae and modulating the activity of multiple signaling molecules at this site. The molecule is synthesized in the endoplasmic reticulum, transported to the cell surface, and undergoes a poorly understood recycling itinerary. We have used mutagenesis to determine the parts of the molecule that control traffic of caveolin-1 from its site of synthesis to the cell surface. We identified four regions of the molecule that appear to influence caveolin-1 traffic. A region between amino acids 66 and 70, which is in the most conserved region of the molecule, is necessary for exit from the endoplasmic reticulum. The region between amino acids 71 and 80 controls incorporation of caveolin-1 oligomers into detergent-resistant regions of the Golgi apparatus. Amino acids 91-100 and 134-154 both control oligomerization and exit from the Golgi apparatus. Removal of other portions of the molecule has no effect on targeting of newly synthesized caveolin-1 to caveolae. The results suggest that movement of caveolin-1 among various endomembrane compartments is controlled at multiple steps.
Figures
Figure 1
Schematic representation of normal and mutant caveolin-1. The 17 different constructs of caveolin-1 used in this study are depicted as bar diagrams, beginning with wild-type caveolin-1 (Cav1-178). Three landmarks in the protein are: the highly conserved signature domain, aa 68–75 (solid box); the scaffolding domain, aa 80–100 (hatched box); and the membrane insertion domain, aa 101–133 (stippled box). The thin connecting line indicates the position of the deleted segment. Eight of the constructs (Cav61A65 to Cav96A100) have the indicated aa replaced with alanine residues. The location of the alanine substitution in each construct is indicated by the gray box. The columns beside each bar diagram indicates the principal site of accumulation in CHO cells of each construct and whether or not it is soluble in Triton X-100.
Figure 2
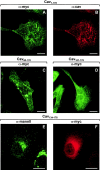
Location of NH2-terminal deletion mutants of caveolin-1. CHO cells either stably transfected with Myc-tagged forms of Cav1-178 (A and B), Cav60-178 (C), or Cav101-178 (D) under control of an IPTG inducible promoter or transiently transfected with Cav134-178 (E and F) were grown on coverslips. Cells were either induced for 16 h with 5 mM IPTG (A–D) or not treated (E and F). CHO cells expressing Cav1-178 were costained with Myc pAb (A) and caveolin mAb (B). CHO cells expressing either Cav60-178 (C) or Cav101-178 (D) were stained with Myc pAb. CHO cells transiently expressing Cav134-178 were double-stained with mannosidase II pAb (E) and Myc mAb (F). Bars, 10 μm.
Figure 3
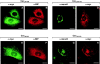
Location of caveolin-1 with COOH-terminal deletions (A and B) and of the membrane insertion domain alone (C). (A and B) CHO cells stably transfected with Myc-tagged Cav1-134 were grown on coverslips and induced for 16 h with 5 mM IPTG. Cells were processed for double immunofluorescence labeling with mannosidase II pAb (A) and Myc mAb (B). (C) CHO cells were transiently transfected with Myc-tagged Cav101-134 and grown on coverslips for 24 h. Cells were processed for indirect immunofluorescence using Myc pAb. Bars, 10 μm.
Figure 4
Effect of internal deletions on location of caveolin-1. CHO cells stably transfected with either Myc-tagged CavΔ60-100 (A and B), CavΔ134-154 (C and D), CavΔ60-80 (E and F), or CavΔ80-100 (G and H) under control of an IPTG inducible promoter were grown on coverslips. Cells were processed for indirect immunofluorescence localization of either the Myc epitope (A, D, E, and H), BIP (B and F), or mannosidase II (C and G). Bars, 10 μm.
Figure 5
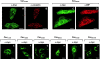
Effect of alanine-substitution on location of caveolin-1. CHO cells stably transfected with either Myc-tagged Cav61A65 (A and B), Cav66A70 (C and D), Cav71A75 (E), Cav76A80 (F), Cav81A85 (G), Cav86A90 (H), Cav91A95 (I), or Cav96A100 (J) under control of an IPTG inducible promoter were grown on coverslips in the presence of IPTG for 16 h. Cells were processed for indirect immunofluorescence localization of the Myc epitope (A, C, and E–J), caveolin-1 (B), or BIP (D). Bars, 10 μm.
Figure 6
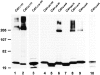
All mutant forms have a wild-type orientation. CHO cells expressing the indicated Myc-tagged caveolin-1 construct were homogenized as described in Materials and Methods. Aliquots of the homogenate were either not treated (lanes 1, 4, 7, 10, 13, 16, and 19), incubated in the presence of 300 μg/ml trypsin for 30 min on ice (lanes 2, 5, 8, 11, 14, 17, and 20), or incubated in the presence of 1% Triton X-100 plus trypsin (lanes 3, 6, 9, 12, 15, 18, and 21). Samples were separated by SDS-PAGE and immunoblotted with Myc pAb.
Figure 7
Turnover of wild-type and mutant caveolin-1. CHO cells stably expressing the indicated Myc-tagged caveolin-1 construct were either not treated (lane 1) or incubated in the presence of 500 μM cycloheximide for 1 (lane 2), 2 (lane 3), 4 (lane 4), 6 (lane 5), or 8 h (lane 6). At the end of the incubation, the cells were lysed, separated by SDS-PAGE, and immunoblotted with Myc pAb. The table indicates the predominant location of the construct as determined by immunofluorescence.
Figure 8
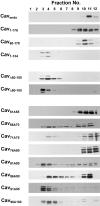
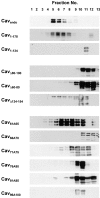
Ability of wild-type and mutant caveolin-1 to form high molecular complexes. CHO cells or CHO cells stably expressing the indicated Myc-tagged caveolin-1 construct were extracted with buffer containing 60 mM octylglucoside. The lysate was loaded on the top of a 5–30% continuous sucrose gradient and centrifuged for 16 h at 340,000 g. Fractions were collected from the top of the gradient, separated by gel electrophoresis, and immunoblotted with either a caveolin-1 pAb (Cavendo) or Myc pAb (Cav1-178 to Cav95A100).
Figure 9
Ability of wild-type and mutant caveolin-1 to form oligomers. Samples of CHO cells expressing the indicated Myc-tagged caveolin construct were collected in sample buffer containing 2% (wt/vol) SDS and 5% (vol/vol) 2-β-mercaptoethanol and solubilized by sonication. Samples were separated on 3–20% polyacrylamide gradient gels and immunoblotted with Myc pAb.
Figure 10
Ability of wild-type and mutant caveolin-1 to incorporate into Triton X-100 insoluble, light membranes. CHO cells or CHO cells stably expressing the indicated Myc-tagged caveolin-1 construct were extracted with 1% Triton X-100 at 4°C overlaid with a 10–30% sucrose gradient. The detergent insoluble fraction was separated from the soluble fraction by centrifugation at 29,000 rpm for 21 h. The gradient was fractionated from the top (fraction 1). Equal volumes of each fraction were separated by SDS-PAGE and immunoblotted with either caveolin-1 pAb (Cavendo) or Myc pAb (Cav1-178 to Cav96A100) as described.
Similar articles
- Caveolin moves from caveolae to the Golgi apparatus in response to cholesterol oxidation.
Smart EJ, Ying YS, Conrad PA, Anderson RG. Smart EJ, et al. J Cell Biol. 1994 Dec;127(5):1185-97. doi: 10.1083/jcb.127.5.1185. J Cell Biol. 1994. PMID: 7962084 Free PMC article. - Palmitoylation of caveolin-1 is required for cholesterol binding, chaperone complex formation, and rapid transport of cholesterol to caveolae.
Uittenbogaard A, Smart EJ. Uittenbogaard A, et al. J Biol Chem. 2000 Aug 18;275(33):25595-9. doi: 10.1074/jbc.M003401200. J Biol Chem. 2000. PMID: 10833523 Retracted. - Interaction of a receptor tyrosine kinase, EGF-R, with caveolins. Caveolin binding negatively regulates tyrosine and serine/threonine kinase activities.
Couet J, Sargiacomo M, Lisanti MP. Couet J, et al. J Biol Chem. 1997 Nov 28;272(48):30429-38. doi: 10.1074/jbc.272.48.30429. J Biol Chem. 1997. PMID: 9374534 - Caveolins and cellular cholesterol balance.
Ikonen E, Parton RG. Ikonen E, et al. Traffic. 2000 Mar;1(3):212-7. doi: 10.1034/j.1600-0854.2000.010303.x. Traffic. 2000. PMID: 11208104 Review. - Intracellular cholesterol transport.
Fielding CJ, Fielding PE. Fielding CJ, et al. J Lipid Res. 1997 Aug;38(8):1503-21. J Lipid Res. 1997. PMID: 9300773 Review.
Cited by
- Probing the caveolin-1 P132L mutant: critical insights into its oligomeric behavior and structure.
Rieth MD, Lee J, Glover KJ. Rieth MD, et al. Biochemistry. 2012 May 8;51(18):3911-8. doi: 10.1021/bi3001853. Epub 2012 Apr 25. Biochemistry. 2012. PMID: 22506673 Free PMC article. - Phospho-caveolin-1 mediates integrin-regulated membrane domain internalization.
del Pozo MA, Balasubramanian N, Alderson NB, Kiosses WB, Grande-García A, Anderson RG, Schwartz MA. del Pozo MA, et al. Nat Cell Biol. 2005 Sep;7(9):901-8. doi: 10.1038/ncb1293. Epub 2005 Aug 21. Nat Cell Biol. 2005. PMID: 16113676 Free PMC article. - Inhibitors caveolin-1 and protein kinase G show differential subcellular colocalization with Nitric oxide synthase.
Adebola TJ, Usha R. Adebola TJ, et al. Afr Health Sci. 2011 Dec;11(4):526-34. Afr Health Sci. 2011. PMID: 22649431 Free PMC article. - Caveolin 1 is required for the activation of endothelial nitric oxide synthase in response to 17beta-estradiol.
Sud N, Wiseman DA, Black SM. Sud N, et al. Mol Endocrinol. 2010 Aug;24(8):1637-49. doi: 10.1210/me.2010-0043. Epub 2010 Jul 7. Mol Endocrinol. 2010. PMID: 20610538 Free PMC article. Retracted. - Caveolin-1: an ambiguous partner in cell signalling and cancer.
Quest AF, Gutierrez-Pajares JL, Torres VA. Quest AF, et al. J Cell Mol Med. 2008 Aug;12(4):1130-50. doi: 10.1111/j.1582-4934.2008.00331.x. Epub 2008 Apr 8. J Cell Mol Med. 2008. PMID: 18400052 Free PMC article. Review.
References
- Anderson R.G.W. The caveolae membrane system. Annu. Rev. Biochem. 1998;67:199–225. - PubMed
- Brewer C.B. Cytomegalovirus plasmid vectors for permanent lines of polarized epithelial cells. Methods Cell Biol. 1994;43:233–245. - PubMed
- Brown D.A., Rose J.K. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. - PubMed
- Das K., Lewis R.Y., Scherer P.E., Lisanti M.P. The membrane-spanning domains of caveolins-1 and -2 mediate the formation of caveolin hetero-oligomers. Implications for the assembly of caveolae membranes in vivo. J. Biol. Chem. 1999;274:18721–18728. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous