A general test of association for quantitative traits in nuclear families - PubMed (original) (raw)
A general test of association for quantitative traits in nuclear families
G R Abecasis et al. Am J Hum Genet. 2000 Jan.
Abstract
High-resolution mapping is an important step in the identification of complex disease genes. In outbred populations, linkage disequilibrium is expected to operate over short distances and could provide a powerful fine-mapping tool. Here we build on recently developed methods for linkage-disequilibrium mapping of quantitative traits to construct a general approach that can accommodate nuclear families of any size, with or without parental information. Variance components are used to construct a test that utilizes information from all available offspring but that is not biased in the presence of linkage or familiality. A permutation test is described for situations in which maximum-likelihood estimates of the variance components are biased. Simulation studies are used to investigate power and error rates of this approach and to highlight situations in which violations of multivariate normality assumptions warrant the permutation test. The relationship between power and the level of linkage disequilibrium for this test suggests that the method is well suited to the analysis of dense maps. The relationship between power and family structure is investigated, and these results are applicable to study design in complex disease, especially for late-onset conditions for which parents are usually not available. When parental genotypes are available, power does not depend greatly on the number of offspring in each family. Power decreases when parental genotypes are not available, but the loss in power is negligible when four or more offspring per family are genotyped. Finally, it is shown that, when siblings are available, the total number of genotypes required in order to achieve comparable power is smaller if parents are not genotyped.
Figures
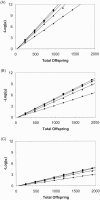
Figure 1
Sensitivity of the orthogonal test to association. Sensitivity is defined as, where α is the significance level exceeded by 80% of simulated data sets. The total number of offspring varied between 240 and 1,920 (in increments of 240 children). Results were plotted for sib-pair families in which parental genotypes were available for analysis (squares) and for sib-pair (diamonds), sib-triad (triangles), and sib-quad (circles) families in which parental genotypes were not available for analysis. The proportion of phenotypic variance attributable to residual sibling resemblance (s_2) was .30. The major-gene effect (h_2) was .10 in panel_A, .05 in panel B, and .025 in panel_C, Each plotted data point corresponds to 1,000 simulated data sets. For convenience, a least-squares straight line has been plotted through each set of data points.
Similar articles
- A method for identifying genes related to a quantitative trait, incorporating multiple siblings and missing parents.
Kistner EO, Weinberg CR. Kistner EO, et al. Genet Epidemiol. 2005 Sep;29(2):155-65. doi: 10.1002/gepi.20084. Genet Epidemiol. 2005. PMID: 16025442 - A sib-pair regression model of linkage disequilibrium for quantitative traits.
Cardon LR. Cardon LR. Hum Hered. 2000 Nov-Dec;50(6):350-8. doi: 10.1159/000022940. Hum Hered. 2000. PMID: 10899753 - The quantitative trait linkage disequilibrium test: a more powerful alternative to the quantitative transmission disequilibrium test for use in the absence of population stratification.
Havill LM, Dyer TD, Richardson DK, Mahaney MC, Blangero J. Havill LM, et al. BMC Genet. 2005 Dec 30;6 Suppl 1(Suppl 1):S91. doi: 10.1186/1471-2156-6-S1-S91. BMC Genet. 2005. PMID: 16451707 Free PMC article. - Genetic linkage methods for quantitative traits.
Amos CI, de Andrade M. Amos CI, et al. Stat Methods Med Res. 2001 Feb;10(1):3-25. doi: 10.1177/096228020101000102. Stat Methods Med Res. 2001. PMID: 11329691 Review. - Joint tests of linkage and association for quantitative traits.
Allison DB, Neale MC. Allison DB, et al. Theor Popul Biol. 2001 Nov;60(3):239-51. doi: 10.1006/tpbi.2001.1544. Theor Popul Biol. 2001. PMID: 11855958 Review.
Cited by
- A family-based study of genetic and epigenetic effects across multiple neurocognitive, motor, social-cognitive and social-behavioral functions.
Nudel R, Zetterberg R, Hemager N, Christiani CAJ, Ohland J, Burton BK, Greve AN, Spang KS, Ellersgaard D, Gantriis DL, Bybjerg-Grauholm J, Plessen KJ, Jepsen JRM, Thorup AAE, Werge T, Mors O, Nordentoft M. Nudel R, et al. Behav Brain Funct. 2022 Dec 1;18(1):14. doi: 10.1186/s12993-022-00198-0. Behav Brain Funct. 2022. PMID: 36457050 Free PMC article. - Causal interpretations of family GWAS in the presence of heterogeneous effects.
Veller C, Przeworski M, Coop G. Veller C, et al. Proc Natl Acad Sci U S A. 2024 Sep 17;121(38):e2401379121. doi: 10.1073/pnas.2401379121. Epub 2024 Sep 13. Proc Natl Acad Sci U S A. 2024. PMID: 39269774 - A general framework for two-stage analysis of genome-wide association studies and its application to case-control studies.
Wason JM, Dudbridge F. Wason JM, et al. Am J Hum Genet. 2012 May 4;90(5):760-73. doi: 10.1016/j.ajhg.2012.03.007. Am J Hum Genet. 2012. PMID: 22560088 Free PMC article. - Association of regions on chromosomes 6 and 7 with blood pressure in Nigerian families.
Tayo BO, Luke A, Zhu X, Adeyemo A, Cooper RS. Tayo BO, et al. Circ Cardiovasc Genet. 2009 Feb;2(1):38-45. doi: 10.1161/CIRCGENETICS.108.817064. Epub 2009 Jan 23. Circ Cardiovasc Genet. 2009. PMID: 20031566 Free PMC article. - TCF7L2 is associated with high serum triacylglycerol and differentially expressed in adipose tissue in families with familial combined hyperlipidaemia.
Huertas-Vazquez A, Plaisier C, Weissglas-Volkov D, Sinsheimer J, Canizales-Quinteros S, Cruz-Bautista I, Nikkola E, Herrera-Hernandez M, Davila-Cervantes A, Tusie-Luna T, Taskinen MR, Aguilar-Salinas C, Pajukanta P. Huertas-Vazquez A, et al. Diabetologia. 2008 Jan;51(1):62-9. doi: 10.1007/s00125-007-0850-6. Epub 2007 Oct 31. Diabetologia. 2008. PMID: 17972059
References
Electronic-Database Information
- The Wellcome Trust Centre for Human Genetics, http://well.ox.ac.uk (for QTDT computer program)
References
- Amos CI, Zhu DK, Boerwinkle E (1996) Assessing genetic linkage and association with robust components of variance approaches. Ann Hum Genet 60:143–160 - PubMed
- Cardon LR. A sib pair regression model of linkage disequilibrium for quantitative traits. Hum Hered (in press) - PubMed
- Chakravarti A (1998) It's raining SNPs, hallelujah? Nat Genet 19:216–217 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources