The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another - PubMed (original) (raw)
The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another
A Das et al. J Bacteriol. 2000 Feb.
Abstract
The VirB proteins of Agrobacterium tumefaciens form a transport pore to transfer DNA from bacteria to plants. The assembly of the transport pore will require interaction among the constituent proteins. The identification of proteins that interact with one another can provide clues to the assembly of the transport pore. We studied interaction among four putative transport pore proteins, VirB7, VirB8, VirB9 and VirB10. Using the yeast two-hybrid assay, we observed that VirB8, VirB9, and VirB10 interact with one another. In vitro studies using protein fusions demonstrated that VirB10 interacts with VirB9 and itself. These results suggest that the outer membrane VirB7-VirB9 complex interacts with the inner membrane proteins VirB8 and VirB10 for the assembly of the transport pore. Fusions that contain small, defined segments of the proteins were used to define the interaction domains of VirB8 and VirB9. All interaction domains of both proteins mapped to the N-terminal half of the proteins. Two separate domains at the N- and C-terminal ends of VirB9 are involved in its homotypic interaction, suggesting that VirB9 forms a higher oligomer. We observed that the alteration of serine at position 87 of VirB8 to leucine abolished its DNA transfer function. Studies on the interaction of the mutant protein with the other VirB proteins showed that the VirB8S87L mutant is defective in interaction with VirB9. The mutant, however, interacted efficiently with VirB8 and VirB10, suggesting that the VirB8-VirB9 interaction is essential for DNA transfer.
Figures
FIG. 1
Interactions of VirB7, VirB8, VirB9, and VirB10. Interaction of VirB proteins was studied by the yeast two-hybrid assay (16). Yeast strains harboring the appropriate plasmids were streaked on solid medium containing X-Gal. A positive interaction is manifested by the blue colony color phenotype. The interactions of VirB7 (B7), VirB8 (B8), VirB9 (B9), and VirB10 (B10) with one another were investigated. One interacting protein is indicated on the left column and the partner proteins are indicated under the colony. For control (C) experiments, the activator vector was introduced into a strain containing the LexA-VirB8, -VirB9, or -VirB10 fusion.
FIG. 2
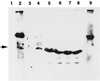
In vitro analysis of the interactions of VirB10. Interaction of VirB10 with VirB8, VirB9, and VirB10 was monitored by the GST-pulldown assay (2). Purified histidine-tagged VirB10 was incubated with GST or a GST fusion protein immobilized on glutathione-Sepharose. Binding of His-VirB10 was determined by analysis of bound proteins by SDS-polyacrylamide gel electrophoresis, followed by Western blot analysis using anti-VirB10 antibodies. Lanes 1 to 4, protein bound to immobilized GST, GST-VirB10, GST-VirB8, and GST-VirB9, respectively; lanes 5 to 8, the corresponding unbound proteins; lane 9, immobilized GST-VirB10 incubated with buffer only. The higher-molecular-weight band in lane 2 is the GST-VirB10 fusion protein. The His-VirB10 protein is indicated by the arrowhead.
FIG. 3
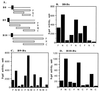
Delineation of interaction domains of VirB8, VirB9, and VirB10. Interactions of VirB8 (B), VirB9 (C), VirB10 (D), and their derivatives with VirB8, VirB9, and VirB10 were investigated by the two-hybrid assay. (A) A linear representation of the VirB proteins and their derivatives used for fusion construction. Fusions containing the periplasmic domain (F), an N-terminal fragment (N), a central fragment (M), and a C-terminal fragment (C) were tested for interaction with the three proteins. The solid box indicates a hydrophobic region.
FIG. 4
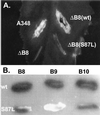
Phenotype of a virB8 mutant. The effect of alteration of VirB8 serine 87 to leucine on DNA transfer was monitored by complementation assays (A). Agrobacterium sp. strain A348ΔB8 (ΔB8) harboring plasmid pAD1433 containing wild-type (wt) virB8 or its derivative pAD1433S87L containing virB8S87L (S87L) was used to infect Kalanchöe leaves. Tumor formation was monitored 3 weeks after infection. A348, Agrobacterium sp. strain A348 that harbors the octopine Ti-plasmid pTiA6. (B) Interaction of VirB8S87L with VirB8, VirB9, and VirB10 was studied by the yeast two-hybrid assays. A blue colony color phenotype indicates a positive interaction. Row 1, VirB8; row 2, VirB8S87L.
FIG. 5
A model for the T-DNA transport pore. Individual VirB proteins are identified by numbers. The T pilus composed of VirB2 can form a channel through the outer membrane.
Similar articles
- Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins: VirB8 is essential for the assembly of the transport pore.
Kumar RB, Xie YH, Das A. Kumar RB, et al. Mol Microbiol. 2000 May;36(3):608-17. doi: 10.1046/j.1365-2958.2000.01876.x. Mol Microbiol. 2000. PMID: 10844650 - Functional analysis of the Agrobacterium tumefaciens T-DNA transport pore protein VirB8.
Kumar RB, Das A. Kumar RB, et al. J Bacteriol. 2001 Jun;183(12):3636-41. doi: 10.1128/JB.183.12.3636-3641.2001. J Bacteriol. 2001. PMID: 11371528 Free PMC article. - Interactions of VirB9, -10, and -11 with the membrane fraction of Agrobacterium tumefaciens: solubility studies provide evidence for tight associations.
Finberg KE, Muth TR, Young SP, Maken JB, Heitritter SM, Binns AN, Banta LM. Finberg KE, et al. J Bacteriol. 1995 Sep;177(17):4881-9. doi: 10.1128/jb.177.17.4881-4889.1995. J Bacteriol. 1995. PMID: 7665464 Free PMC article. - VirB8: a conserved type IV secretion system assembly factor and drug target.
Baron C. Baron C. Biochem Cell Biol. 2006 Dec;84(6):890-9. doi: 10.1139/o06-148. Biochem Cell Biol. 2006. PMID: 17215876 Review. - Assembly of the VirB transport complex for DNA transfer from Agrobacterium tumefaciens to plant cells.
Zupan JR, Ward D, Zambryski P. Zupan JR, et al. Curr Opin Microbiol. 1998 Dec;1(6):649-55. doi: 10.1016/s1369-5274(98)80110-0. Curr Opin Microbiol. 1998. PMID: 10066547 Review.
Cited by
- Complete nucleotide sequence of the conjugative tetracycline resistance plasmid pFBAOT6, a member of a group of IncU plasmids with global ubiquity.
Rhodes G, Parkhill J, Bird C, Ambrose K, Jones MC, Huys G, Swings J, Pickup RW. Rhodes G, et al. Appl Environ Microbiol. 2004 Dec;70(12):7497-510. doi: 10.1128/AEM.70.12.7497-7510.2004. Appl Environ Microbiol. 2004. PMID: 15574953 Free PMC article. - Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis.
Jakubowski SJ, Kerr JE, Garza I, Krishnamoorthy V, Bayliss R, Waksman G, Christie PJ. Jakubowski SJ, et al. Mol Microbiol. 2009 Feb;71(3):779-94. doi: 10.1111/j.1365-2958.2008.06565.x. Epub 2008 Dec 1. Mol Microbiol. 2009. PMID: 19054325 Free PMC article. - An in vivo high-throughput screening approach targeting the type IV secretion system component VirB8 identified inhibitors of Brucella abortus 2308 proliferation.
Paschos A, den Hartigh A, Smith MA, Atluri VL, Sivanesan D, Tsolis RM, Baron C. Paschos A, et al. Infect Immun. 2011 Mar;79(3):1033-43. doi: 10.1128/IAI.00993-10. Epub 2010 Dec 20. Infect Immun. 2011. PMID: 21173315 Free PMC article. - Structural Insight into How Bacteria Prevent Interference between Multiple Divergent Type IV Secretion Systems.
Gillespie JJ, Phan IQ, Scheib H, Subramanian S, Edwards TE, Lehman SS, Piitulainen H, Rahman MS, Rennoll-Bankert KE, Staker BL, Taira S, Stacy R, Myler PJ, Azad AF, Pulliainen AT. Gillespie JJ, et al. mBio. 2015 Dec 8;6(6):e01867-15. doi: 10.1128/mBio.01867-15. mBio. 2015. PMID: 26646013 Free PMC article. - Protein interaction mapping on a functional shotgun sequence of Rickettsia sibirica.
Malek JA, Wierzbowski JM, Tao W, Bosak SA, Saranga DJ, Doucette-Stamm L, Smith DR, McEwan PJ, McKernan KJ. Malek JA, et al. Nucleic Acids Res. 2004 Feb 10;32(3):1059-64. doi: 10.1093/nar/gkh254. Print 2004. Nucleic Acids Res. 2004. PMID: 14872061 Free PMC article.
References
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene and Wiley-Interscience; 1993.
- Beijersbergen A, Dulk-Ras A D, Schilperoort R A, Hooykaas P J. Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science. 1992;256:1324–1327. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases